Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site
- PMID: 16204552
- PMCID: PMC1265972
- DOI: 10.1128/AEM.71.10.6308-6318.2005
Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site
Abstract
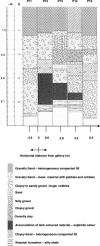
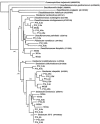
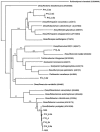
The geochemistry and microbiology of a uranium-contaminated subsurface environment that had undergone two seasons of acetate addition to stimulate microbial U(VI) reduction was examined. There were distinct horizontal and vertical geochemical gradients that could be attributed in large part to the manner in which acetate was distributed in the aquifer, with more reduction of Fe(III) and sulfate occurring at greater depths and closer to the point of acetate injection. Clone libraries of 16S rRNA genes derived from sediments and groundwater indicated an enrichment of sulfate-reducing bacteria in the order Desulfobacterales in sediment and groundwater samples. These samples were collected nearest the injection gallery where microbially reducible Fe(III) oxides were highly depleted, groundwater sulfate concentrations were low, and increases in acid volatile sulfide were observed in the sediment. Further down-gradient, metal-reducing conditions were present as indicated by intermediate Fe(II)/Fe(total) ratios, lower acid volatile sulfide values, and increased abundance of 16S rRNA gene sequences belonging to the dissimilatory Fe(III)- and U(VI)-reducing family Geobacteraceae. Maximal Fe(III) and U(VI) reduction correlated with maximal recovery of Geobacteraceae 16S rRNA gene sequences in both groundwater and sediment; however, the sites at which these maxima occurred were spatially separated within the aquifer. The substantial microbial and geochemical heterogeneity at this site demonstrates that attempts should be made to deliver acetate in a more uniform manner and that closely spaced sampling intervals, horizontally and vertically, in both sediment and groundwater are necessary in order to obtain a more in-depth understanding of microbial processes and the relative contribution of attached and planktonic populations to in situ uranium bioremediation.
Figures



Similar articles
-
Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI).Appl Environ Microbiol. 2003 Dec;69(12):7467-79. doi: 10.1128/AEM.69.12.7467-7479.2003. Appl Environ Microbiol. 2003. PMID: 14660400 Free PMC article.
-
Microorganisms associated with uranium bioremediation in a high-salinity subsurface sediment.Appl Environ Microbiol. 2003 Jun;69(6):3672-5. doi: 10.1128/AEM.69.6.3672-3675.2003. Appl Environ Microbiol. 2003. PMID: 12788780 Free PMC article.
-
Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate.Appl Environ Microbiol. 2004 Aug;70(8):4911-20. doi: 10.1128/AEM.70.8.4911-4920.2004. Appl Environ Microbiol. 2004. PMID: 15294831 Free PMC article.
-
Bioremediation of uranium-contaminated groundwater: a systems approach to subsurface biogeochemistry.Curr Opin Biotechnol. 2013 Jun;24(3):489-97. doi: 10.1016/j.copbio.2012.10.008. Epub 2012 Nov 16. Curr Opin Biotechnol. 2013. PMID: 23159488 Review.
-
Extracellular electron transfer-dependent bioremediation of uranium-contaminated groundwater: Advancements and challenges.Water Res. 2025 Mar 15;272:122957. doi: 10.1016/j.watres.2024.122957. Epub 2024 Dec 12. Water Res. 2025. PMID: 39708382 Review.
Cited by
-
Spatial heterogeneity of bacterial communities in sediments from an infiltration basin receiving highway runoff.Microb Ecol. 2012 Aug;64(2):461-73. doi: 10.1007/s00248-012-0026-x. Epub 2012 Mar 6. Microb Ecol. 2012. PMID: 22391798
-
Profiling in situ microbial community structure with an amplification microarray.Appl Environ Microbiol. 2013 Feb;79(3):799-807. doi: 10.1128/AEM.02664-12. Epub 2012 Nov 16. Appl Environ Microbiol. 2013. PMID: 23160129 Free PMC article.
-
Genome-wide gene expression patterns and growth requirements suggest that Pelobacter carbinolicus reduces Fe(III) indirectly via sulfide production.Appl Environ Microbiol. 2008 Jul;74(14):4277-84. doi: 10.1128/AEM.02901-07. Epub 2008 May 30. Appl Environ Microbiol. 2008. PMID: 18515480 Free PMC article.
-
Constraint-based modeling analysis of the metabolism of two Pelobacter species.BMC Syst Biol. 2010 Dec 23;4:174. doi: 10.1186/1752-0509-4-174. BMC Syst Biol. 2010. PMID: 21182788 Free PMC article.
-
Meta-Transcriptomes From Microcosms From a Cr Impacted Soil Provides Insights Into the Metabolic Response of the Microbial Populations to Acetate Stimulation.Environ Microbiol Rep. 2025 Aug;17(4):e70148. doi: 10.1111/1758-2229.70148. Environ Microbiol Rep. 2025. PMID: 40624790 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. - PMC - PubMed
-
- Chapelle, F. H., and D. R. Lovley. 1992. Competitive exclusion of sulphate reduction by Fe(III)-reducing bacteria: a mechanism for producing discrete zones of high-iron groundwater. Ground Water 30:29-36.
-
- Cline, J. D. 1969. Spectrophotmetric determination of hydrogen sulphide in natural waters. Limnol. Oceanogr. 14:454-458.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

