Positive selection of a pre-expansion CAG repeat of the human SCA2 gene
- PMID: 16205789
- PMCID: PMC1239938
- DOI: 10.1371/journal.pgen.0010041
Positive selection of a pre-expansion CAG repeat of the human SCA2 gene
Abstract
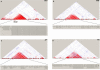
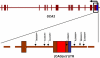
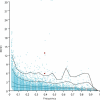
A region of approximately one megabase of human Chromosome 12 shows extensive linkage disequilibrium in Utah residents with ancestry from northern and western Europe. This strikingly large linkage disequilibrium block was analyzed with statistical and experimental methods to determine whether natural selection could be implicated in shaping the current genome structure. Extended Haplotype Homozygosity and Relative Extended Haplotype Homozygosity analyses on this region mapped a core region of the strongest conserved haplotype to the exon 1 of the Spinocerebellar ataxia type 2 gene (SCA2). Direct DNA sequencing of this region of the SCA2 gene revealed a significant association between a pre-expanded allele [(CAG)8CAA(CAG)4CAA(CAG)8] of CAG repeats within exon 1 and the selected haplotype of the SCA2 gene. A significantly negative Tajima's D value (-2.20, p < 0.01) on this site consistently suggested selection on the CAG repeat. This region was also investigated in the three other populations, none of which showed signs of selection. These results suggest that a recent positive selection of the pre-expansion SCA2 CAG repeat has occurred in Utah residents with European ancestry.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
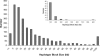


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

