Histone structure and nucleosome stability
- PMID: 16209651
- PMCID: PMC1831843
- DOI: 10.1586/14789450.2.5.719
Histone structure and nucleosome stability
Abstract
Histone proteins play essential structural and functional roles in the transition between active and inactive chromatin states. Although histones have a high degree of conservation due to constraints to maintain the overall structure of the nucleosomal octameric core, variants have evolved to assume diverse roles in gene regulation and epigenetic silencing. Histone variants, post-translational modifications and interactions with chromatin remodeling complexes influence DNA replication, transcription, repair and recombination. The authors review recent findings on the structure of chromatin that confirm previous interparticle interactions observed in crystal structures.
Figures
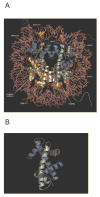



References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389(6648):251–260. - PubMed
-
•• Contains high-resolution structures of the nucleosome that are key to understanding gene regulation and chromatin remodeling.
-
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002 ;319(5):1097–1113. - PubMed
-
•• Contains high-resolution structures of the nucleosome that are key to understanding gene regulation and chromatin remodeling.
-
- Dorigo B, Schalch T, Kulangara A, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306(5701):1571–1573. - PubMed
-
• Provides evidence for a two-start helical organization of the 30-nm chromatin fiber and has helped resolve a controversy over how individual nucleosomes are packed in the chromatin fiber.
-
- van Holde KE. Chromatin. Springer-Verlag; London, UK: 1988. Chapter 7: Highter order structure; pp. 317–354.
-
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306(5701):1574–1577. - PubMed
Websites
-
- Marino-Ramirez L, Hsu B, Baxevanis AD, Landsman D NHGRI/NCBI. http://research.nhgri.nih.gov/histones (Viewed September 2005)
-
- Delano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. http://www.pymol.org (Viewed September 2005)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
