Microcoding: the second step in DNA barcoding
- PMID: 16214747
- PMCID: PMC1609221
- DOI: 10.1098/rstb.2005.1721
Microcoding: the second step in DNA barcoding
Abstract
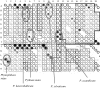
After the process of DNA barcoding has become well advanced in a group of organisms, as it has in the economically important fungi, the question then arises as to whether shorter and literally more barcode-like DNA segments should be utilized to facilitate rapid identification and, where applicable, detection. Through appropriate software analysis of typical full-length barcodes (generally over 500 base pairs long), uniquely distinctive oligonucleotide 'microcodes' of less than 25 bp can be found that allow rapid identification of circa 100-200 species on various array-like platforms. Microarrays can in principle fulfill the function of microcode-based species identification but, because of their high cost and low level of reusability, they tend to be less cost-effective. Two alternative platforms in current use in fungal identification are reusable nylon-based macroarrays and the Luminex system of specific, colour-coded DNA detection beads analysed by means of a flow cytometer. When the most efficient means of rapid barcode-based species identification is sought, a choice can be made either for one of these methodologies or for basic high-throughput sequencing, depending on the strategic outlook of the investigator and on current costs. Arrays and functionally similar platforms may have a particular advantage when a biologically complex material such as soil or a human respiratory secretion sample is analysed to give a census of relevant species present.
Figures
References
-
- Aanen D.K, Kuyper T.W, Mes T.H, Hoekstra R.F. The evolution of reproductive isolation in the ectomycorrhizal Hebeloma crustuliniforme aggregate (Basidiomycetes) in northwestern Europe: a phylogenetic approach. Evol. Int. J. Org. Evol. 2000;54:1192–1206. - PubMed
-
- Anonymous . Top Crop Manager. 2002. Early season diseases take their toll. Dec. 2002, http://www.topcropmanager.com/6_search/article_e.asp?article=1300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
