Simulation study of the contribution of oligomer/oligomer binding to capsid assembly kinetics
- PMID: 16214864
- PMCID: PMC1367037
- DOI: 10.1529/biophysj.105.072207
Simulation study of the contribution of oligomer/oligomer binding to capsid assembly kinetics
Abstract
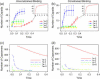
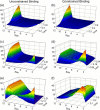
The process by which hundreds of identical capsid proteins self-assemble into icosahedral structures is complex and poorly understood. Establishing constraints on the assembly pathways is crucial to building reliable theoretical models. For example, it is currently an open question to what degree overall assembly kinetics are dominated by one or a few most efficient pathways versus the enormous number theoretically possible. The importance of this question, however, is often overlooked due to the difficulties of addressing it in either theoretical or experimental practice. We apply a computer model based on a discrete-event simulation method to evaluate the contributions of nondominant pathways to overall assembly kinetics. This is accomplished by comparing two possible assembly models: one allowing growth to proceed only by the accretion of individual assembly subunits and the other allowing the binding of sterically compatible assembly intermediates any sizes. Simulations show that the two models perform almost identically under low binding rate conditions, where growth is strongly nucleation-limited, but sharply diverge under conditions of higher association rates or coat protein concentrations. The results suggest the importance of identifying the actual binding pattern if one is to build reliable models of capsid assembly or other complex self-assembly processes.
Figures




Similar articles
-
Kinetic theory of virus capsid assembly.Phys Biol. 2007 Nov 26;4(4):296-304. doi: 10.1088/1478-3975/4/4/006. Phys Biol. 2007. PMID: 18185007
-
Local rules simulation of the kinetics of virus capsid self-assembly.Biophys J. 1998 Dec;75(6):2626-36. doi: 10.1016/S0006-3495(98)77708-2. Biophys J. 1998. PMID: 9826587 Free PMC article.
-
Modelling the self-assembly of virus capsids.J Phys Condens Matter. 2010 Mar 17;22(10):104101. doi: 10.1088/0953-8984/22/10/104101. Epub 2010 Feb 23. J Phys Condens Matter. 2010. PMID: 21389435
-
The energy landscape as a unifying theme in molecular science.Philos Trans A Math Phys Eng Sci. 2005 Feb 15;363(1827):357-75; discussion 375-7. doi: 10.1098/rsta.2004.1497. Philos Trans A Math Phys Eng Sci. 2005. PMID: 15664888 Review.
-
Adenovirus complex structures.Curr Opin Struct Biol. 2005 Apr;15(2):237-43. doi: 10.1016/j.sbi.2005.03.004. Curr Opin Struct Biol. 2005. PMID: 15837184 Review.
Cited by
-
Quantitative computational models of molecular self-assembly in systems biology.Phys Biol. 2017 May 23;14(3):035003. doi: 10.1088/1478-3975/aa6cdc. Phys Biol. 2017. PMID: 28535149 Free PMC article. Review.
-
Nanofluidic Devices with 8 Pores in Series for Real-Time, Resistive-Pulse Analysis of Hepatitis B Virus Capsid Assembly.Anal Chem. 2017 May 2;89(9):4855-4862. doi: 10.1021/acs.analchem.6b04491. Epub 2017 Apr 17. Anal Chem. 2017. PMID: 28322548 Free PMC article.
-
Exploring the parameter space of complex self-assembly through virus capsid models.Biophys J. 2008 Feb 1;94(3):772-83. doi: 10.1529/biophysj.107.107284. Epub 2007 Oct 5. Biophys J. 2008. PMID: 17921216 Free PMC article.
-
Stochastic kinetics of viral capsid assembly based on detailed protein structures.Biophys J. 2006 May 1;90(9):3029-42. doi: 10.1529/biophysj.105.076737. Epub 2006 Feb 10. Biophys J. 2006. PMID: 16473916 Free PMC article.
-
Recent advances in coarse-grained modeling of virus assembly.Curr Opin Virol. 2016 Jun;18:36-43. doi: 10.1016/j.coviro.2016.02.012. Epub 2016 Mar 24. Curr Opin Virol. 2016. PMID: 27016708 Free PMC article. Review. No abstract available.
References
-
- Caspar, D. L., and A. Klug. 1962. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27:1–24. - PubMed
-
- Prevelige, P. E., Jr. 1998. Inhibiting virus-capsid assembly by altering the polymerization pathway. Trends Biotechnol. 16:61–65. - PubMed
-
- Zlotnick, A., and S. J. Stray. 2003. How does your virus grow? Understanding and interfering with virus assembly. Trends Biotechnol. 21:536–542. - PubMed
-
- Whitesides, G. M., and B. Grzybowski. 2002. Self-assembly at all scales. Science. 295:2418–2421. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

