Mechanics and dynamics of actin-driven thin membrane protrusions
- PMID: 16214866
- PMCID: PMC1367038
- DOI: 10.1529/biophysj.105.071480
Mechanics and dynamics of actin-driven thin membrane protrusions
Abstract
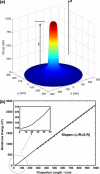
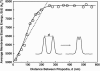
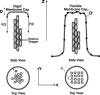
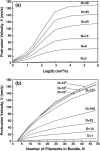
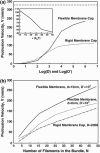
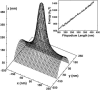
Motile cells explore their surrounding milieu by extending thin dynamic protrusions, or filopodia. The growth of filopodia is driven by actin filament bundles that polymerize underneath the cell membrane. We compute the mechanical and dynamical features of the protrusion growth process by explicitly incorporating the flexible plasma membrane. We find that a critical number of filaments are needed to generate net filopodial growth. Without external influences, the filopodium can extend indefinitely up to the buckling length of the F-actin bundle. Dynamical calculations show that the protrusion speed is enhanced by the thermal fluctuations of the membrane; a filament bundle encased in a flexible membrane grows much faster. The protrusion speed depends directly on the number and spatial arrangement of the filaments in the bundle and whether the filaments are tethered to the membrane. Filopodia also attract each other through distortions of the membrane. Spatially close filopodia will merge to form a larger one. Force-velocity relationships mimicking micromanipulation experiments testing our predictions are computed.
Figures



References
-
- Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filaments dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. - PubMed
-
- Borisy, G. G., and T. M. Svitkina. 2000. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 12:104–112. - PubMed
-
- Small, J. V. 1988. The actin cytoskeleton. Electron Microsc. Rev. 1:155–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

