Probing the mechanical folding kinetics of TAR RNA by hopping, force-jump, and force-ramp methods
- PMID: 16214869
- PMCID: PMC1367024
- DOI: 10.1529/biophysj.105.068049
Probing the mechanical folding kinetics of TAR RNA by hopping, force-jump, and force-ramp methods
Abstract
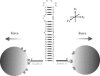
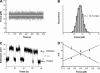
Mechanical unfolding and refolding of single RNA molecules have previously been observed in optical traps as sudden changes in molecular extension. Two methods have been traditionally used: "force-ramp", with the applied force continuously changing, and "hopping". In hopping experiments the force is held constant and the molecule jumps spontaneously between two different states. Unfolding/refolding rates are measured directly, but only over a very narrow range of forces. We have now developed a force-jump method to measure the unfolding and refolding rates independently over a wider range of forces. In this method, the applied force is rapidly stepped to a new value and either the unfolding or refolding event is monitored through changes in the molecular extension. The force-jump technique is compared to the force-ramp and hopping methods by using a 52-nucleotide RNA hairpin with a three-nucleotide bulge, i.e., the transactivation response region RNA from the human immunodeficiency virus. We find the unfolding kinetics and Gibbs free energies obtained from all three methods to be in good agreement. The transactivation response region RNA hairpin unfolds in an all-or-none two-state reaction at any loading rate with the force-ramp method. The unfolding reaction is reversible at small loading rates, but shows hysteresis at higher loading rates. Although the RNA unfolds and refolds without detectable intermediates in constant-force conditions (hopping and force-jump), it shows partially folded intermediates in force-ramp experiments at higher unloading rates. Thus, we find that folding of RNA hairpins can be more complex than a simple single-step reaction, and that application of several methods can improve understanding of reaction mechanisms.
Figures
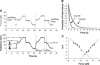



Similar articles
-
Mechanical unfolding of RNA: from hairpins to structures with internal multiloops.Biophys J. 2007 Feb 1;92(3):731-43. doi: 10.1529/biophysj.106.093062. Epub 2006 Oct 6. Biophys J. 2007. PMID: 17028142 Free PMC article.
-
Forced-unfolding and force-quench refolding of RNA hairpins.Biophys J. 2006 May 15;90(10):3410-27. doi: 10.1529/biophysj.105.078030. Epub 2006 Feb 10. Biophys J. 2006. PMID: 16473903 Free PMC article.
-
Monte Carlo simulation for single RNA unfolding by force.Biophys J. 2005 Jan;88(1):76-84. doi: 10.1529/biophysj.104.049239. Epub 2004 Oct 22. Biophys J. 2005. PMID: 15501942 Free PMC article.
-
The effect of force on thermodynamics and kinetics: unfolding single RNA molecules.Biochem Soc Trans. 2004 Nov;32(Pt 5):757-60. doi: 10.1042/BST0320757. Biochem Soc Trans. 2004. PMID: 15494007 Free PMC article. Review.
-
Determination of thermodynamics and kinetics of RNA reactions by force.Q Rev Biophys. 2006 Nov;39(4):325-60. doi: 10.1017/S0033583506004446. Epub 2006 Oct 16. Q Rev Biophys. 2006. PMID: 17040613 Free PMC article. Review.
Cited by
-
Dwell-time distribution analysis of polyprotein unfolding using force-clamp spectroscopy.Biophys J. 2007 Apr 15;92(8):2896-903. doi: 10.1529/biophysj.106.099481. Epub 2007 Jan 26. Biophys J. 2007. PMID: 17259284 Free PMC article.
-
Force Dependence of Proteins' Transition State Position and the Bell-Evans Model.Nanomaterials (Basel). 2021 Nov 11;11(11):3023. doi: 10.3390/nano11113023. Nanomaterials (Basel). 2021. PMID: 34835787 Free PMC article.
-
Biological mechanisms, one molecule at a time.Genes Dev. 2011 Jun 15;25(12):1205-31. doi: 10.1101/gad.2050011. Genes Dev. 2011. PMID: 21685361 Free PMC article. Review.
-
Nanomanipulation of single RNA molecules by optical tweezers.J Vis Exp. 2014 Aug 20;(90):51542. doi: 10.3791/51542. J Vis Exp. 2014. PMID: 25177917 Free PMC article.
-
Optical Tweezers to Study Viruses.Subcell Biochem. 2024;105:359-399. doi: 10.1007/978-3-031-65187-8_10. Subcell Biochem. 2024. PMID: 39738952 Review.
References
-
- Leckband, D. 2000. Measuring the forces that control protein interactions. Annu. Rev. Biophys. Biomol. Struct. 9:1–26. - PubMed
-
- Muller, D., J. Heymann, F. Oesterhelt, C. Moller, H. Gaub, G. Buldt, and A. Engel. 2000. Atomic force microscopy of native purple membrane. Biochim. Biophys. Acta. 1460:27–38. - PubMed
-
- Hugel, T., N. Holland, A. Cattani, L. Moroder, M. Seitz, and H. Gaub. 2002. Single-molecule optomechanical cycle. Science. 296:1103–1106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

