Effects of disulfide bonds on folding behavior and mechanism of the beta-sheet protein tendamistat
- PMID: 16214873
- PMCID: PMC1367026
- DOI: 10.1529/biophysj.105.063552
Effects of disulfide bonds on folding behavior and mechanism of the beta-sheet protein tendamistat
Abstract
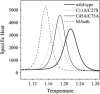
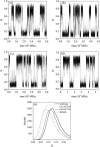
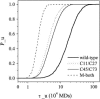
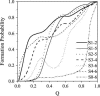
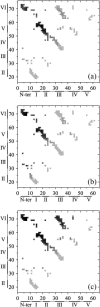
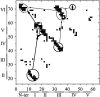
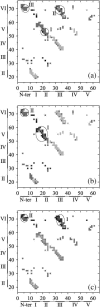
Tendamistat, a small disulfide-bonded beta-sheet protein, and its three single/double-disulfide mutants are investigated by using a modified Gō-like model, aiming to understand the folding mechanism of disulfide-bonded protein as well as the effects of removal of disulfide bond on the folding process. Our simulations show that tendamistat and its two single-disulfide mutants are all two-state folders, consistent with the experimental observations. It is found that the disulfide bonds as well as three hydrogen bonds between the N-terminal loop-0 and strand-6 are of significant importance for the folding of tendamistat. Without these interactions, their two-state behaviors become unstable and the predictions of the model are inconsistent with experiments. In addition, the effect of disulfide bonds on the folding process are studied by comparing the wild-type tendamistat and its two mutants; it is found that the removal of either of the C11-C27 or C45-C73 disulfide bond leads to a large decrease in the thermodynamical stability and loss of structure in the unfolded state, and the effect of the former is stronger than that of the later. These simulation results are in good agreement with experiments and, thus, validate our model. Based on the same model, the detailed folding pathways of the wild-type tendamistat and two mutants are studied, and the effect of disulfide bonds on the folding kinetics are discussed. The obtained results provide a detailed folding picture of these proteins and complement experimental findings. Finally, the folding nuclei predicted to be existent in this protein tendamistat as well as its mutants are firstly identified in this work. The positions of the nucleus are consistent with those argued in experimental studies. Therefore, a nucleation/growth folding mechanism that can explain the two-state folding manner is clearly characterized. Moreover, the effect by the removal of each disulfide bond on the folding thermodynamics and dynamics can also be well interpreted from their influence on the folding nucleus. The implementation of this work indicates that the modified Gō-like model really describes the folding behavior of protein tendamistat and could be used to study the folding of other disulfide-bonded proteins.
Figures
Similar articles
-
Effect of preformed correct tertiary interactions on rapid two-state tendamistat folding: evidence for hairpins as initiation sites for beta-sheet formation.Biochemistry. 1997 Jul 22;36(29):9057-65. doi: 10.1021/bi970594r. Biochemistry. 1997. PMID: 9220995
-
Mechanism of protein stabilization by disulfide bridges: calorimetric unfolding studies on disulfide-deficient mutants of the alpha-amylase inhibitor tendamistat.J Mol Biol. 1995 Dec 1;254(3):481-96. doi: 10.1006/jmbi.1995.0632. J Mol Biol. 1995. PMID: 7490764
-
Folding of the disulfide-bonded beta-sheet protein tendamistat: rapid two-state folding without hydrophobic collapse.J Mol Biol. 1997 May 2;268(2):526-38. doi: 10.1006/jmbi.1997.0960. J Mol Biol. 1997. PMID: 9159488
-
Protein folding simulations: from coarse-grained model to all-atom model.IUBMB Life. 2009 Jun;61(6):627-43. doi: 10.1002/iub.223. IUBMB Life. 2009. PMID: 19472192 Review.
-
Overview of the regulation of disulfide bond formation in Peptide and protein folding.Curr Protoc Protein Sci. 2014 Apr 1;76:28.6.1-28.6.6. doi: 10.1002/0471140864.ps2806s76. Curr Protoc Protein Sci. 2014. PMID: 24692015 Review.
Cited by
-
Analysis of core-periphery organization in protein contact networks reveals groups of structurally and functionally critical residues.J Biosci. 2015 Oct;40(4):683-99. doi: 10.1007/s12038-015-9554-0. J Biosci. 2015. PMID: 26564971
-
Protein folding guides disulfide bond formation.Proc Natl Acad Sci U S A. 2015 Sep 8;112(36):11241-6. doi: 10.1073/pnas.1503909112. Epub 2015 Aug 21. Proc Natl Acad Sci U S A. 2015. PMID: 26297249 Free PMC article.
-
Simple MD-based model for oxidative folding of peptides and proteins.Sci Rep. 2017 Aug 24;7(1):9293. doi: 10.1038/s41598-017-09229-7. Sci Rep. 2017. PMID: 28839177 Free PMC article.
-
A continuous-exchange cell-free protein synthesis system based on extracts from cultured insect cells.PLoS One. 2014 May 7;9(5):e96635. doi: 10.1371/journal.pone.0096635. eCollection 2014. PLoS One. 2014. PMID: 24804975 Free PMC article.
-
Effect of Glycosylation on an Immunodominant Region in the V1V2 Variable Domain of the HIV-1 Envelope gp120 Protein.PLoS Comput Biol. 2016 Oct 7;12(10):e1005094. doi: 10.1371/journal.pcbi.1005094. eCollection 2016 Oct. PLoS Comput Biol. 2016. PMID: 27716795 Free PMC article.
References
-
- Wolynes, P. G., J. N. Onuchic, and D. Thirumalai. 1995. Navigating the folding routes. Science. 267:1619–1620. - PubMed
-
- Onuchic, J. N., and P. G. Wolynes. 2004. Theory of protein folding. Curr. Opin. Struct. Biol. 14:70–75. - PubMed
-
- Anfinsen, C. B. 1973. Principles that govern the folding of protein chains. Science. 181:223–230. - PubMed
-
- Fersht, A. R. 1999. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. Freeman, New York, NY.
-
- Daggett, V., and A. R. Fersht. 2003. The present view of the mechanism of protein folding. Nat. Rev. Mol. Cell Biol. 4:497–502. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

