The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence
- PMID: 16214898
- PMCID: PMC1276025
- DOI: 10.1105/tpc.105.036004
The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence
Abstract
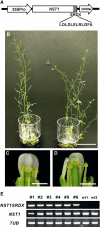
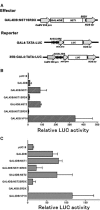
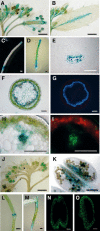
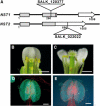
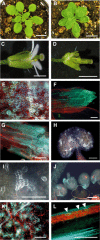
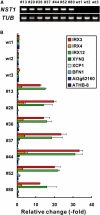
In plants, secondary wall thickenings play important roles in various biological processes, although the factors regulating these processes remain to be characterized. We show that expression of chimeric repressors derived from NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and NST2 in Arabidopsis thaliana resulted in an anther dehiscence defect due to loss of secondary wall thickening in anther endothecium. Plants with double, but not single, T-DNA-tagged lines for NST1 and NST2 had the same anther-indehiscent phenotype as transgenic plants that expressed the individual chimeric repressors, indicating that NST1 and NST2 are redundant in regulating secondary wall thickening in anther walls. The activity of the NST2 promoter was particularly strong in anther tissue, while that of the NST1 promoter was detected in various tissues in which lignified secondary walls develop. Ectopic expression of NST1 or NST2 induced ectopic thickening of secondary walls in various aboveground tissues. Epidermal cells with ectopic thickening of secondary walls had structural features similar to those of tracheary elements. However, among genes involved in the differentiation of tracheary elements, only those related to secondary wall synthesis were clearly upregulated. None of the genes involved in programmed cell death were similarly affected. Our results suggest NAC transcription factors as possible regulators of secondary wall thickening in various tissues.
Figures

References
-
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182. - PubMed
-
- Cano-Delgado, A.I., Metzlaff, K., and Bevan, M.W. (2000). The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127, 3395–3405. - PubMed
NOTE ADDED IN PROOF
-
- While this manuscript was under review, Kubo et al. (2005) described that the NAC domain transcription factors VND6 (At5g62380) and VND7 (At1g71930) induced ectopic metaxylem- and protoxylem-like vessel elements, respectively, in Arabidopsis when overexpressed.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

