Protein database searches using compositionally adjusted substitution matrices
- PMID: 16218944
- PMCID: PMC1343503
- DOI: 10.1111/j.1742-4658.2005.04945.x
Protein database searches using compositionally adjusted substitution matrices
Abstract
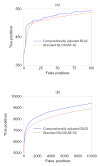
Almost all protein database search methods use amino acid substitution matrices for scoring, optimizing, and assessing the statistical significance of sequence alignments. Much care and effort has therefore gone into constructing substitution matrices, and the quality of search results can depend strongly upon the choice of the proper matrix. A long-standing problem has been the comparison of sequences with biased amino acid compositions, for which standard substitution matrices are not optimal. To address this problem, we have recently developed a general procedure for transforming a standard matrix into one appropriate for the comparison of two sequences with arbitrary, and possibly differing compositions. Such adjusted matrices yield, on average, improved alignments and alignment scores when applied to the comparison of proteins with markedly biased compositions. Here we review the application of compositionally adjusted matrices and consider whether they may also be applied fruitfully to general purpose protein sequence database searches, in which related sequence pairs do not necessarily have strong compositional biases. Although it is not advisable to apply compositional adjustment indiscriminately, we describe several simple criteria under which invoking such adjustment is on average beneficial. In a typical database search, at least one of these criteria is satisfied by over half the related sequence pairs. Compositional substitution matrix adjustment is now available in NCBI's protein-protein version of blast.
Figures
Comment in
-
Identifying protein interactions.FEBS J. 2005 Oct;272(20):5099-100. doi: 10.1111/j.1742-4658.2005.04944.x. FEBS J. 2005. PMID: 16218943 No abstract available.
References
-
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–53. - PubMed
-
- McLachlan AD. Tests for comparing related amino-acid sequences. Cytochrome c and cytochrome c 551. J Mol Biol. 1971;61:409–24. - PubMed
-
- Dayhoff, M. O., Schwartz, R. M. & Orcutt, B. C. (1978) A model of evolutionary change in proteins in Atlas of Protein Sequence and Structure (Dayhoff, M. O., ed) pp. 345–52, Natl Biomed Res Found, Washington, DC.
-
- Schwartz, R. M. & Dayhoff, M. O. (1978) Matrices for detecting distant relationships in Atlas of Protein Sequence and Structure (Dayhoff, M. O., ed) pp. 353–58, Natl Biomed Res Found, Washington, DC.
-
- Feng DF, Johnson MS, Doolittle RF. Aligning amino acid sequences: comparison of commonly used methods. J Mol Evol. 1984;21:112–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

