Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs
- PMID: 16218957
- PMCID: PMC1360291
- DOI: 10.1111/j.1742-4658.2005.04923.x
Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs
Abstract
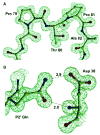
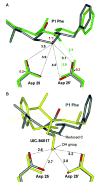
HIV-1 protease (PR) and two drug-resistant variants--PR with the V82A mutation (PR(V82A)) and PR with the I84V mutation (PR(I84V))--were studied using reduced peptide analogs of five natural cleavage sites (CA-p2, p2-NC, p6pol-PR, p1-p6 and NC-p1) to understand the structural and kinetic changes. The common drug-resistant mutations V82A and I84V alter residues forming the substrate-binding site. Eight crystal structures were refined at resolutions of 1.10-1.60 A. Differences in the PR-analog interactions depended on the peptide sequence and were consistent with the relative inhibition. Analog p6(pol)-PR formed more hydrogen bonds of P2 Asn with PR and fewer van der Waals contacts at P1' Pro compared with those formed by CA-p2 or p2-NC in PR complexes. The P3 Gly in p1-p6 provided fewer van der Waals contacts and hydrogen bonds at P2-P3 and more water-mediated interactions. PR(I84V) showed reduced van der Waals interactions with inhibitor compared with PR, which was consistent with kinetic data. The structures suggest that the binding affinity for mutants is modulated by the conformational flexibility of the substrate analogs. The complexes of PR(V82A) showed smaller shifts of the main chain atoms of Ala82 relative to PR, but more movement of the peptide analog, compared to complexes with clinical inhibitors. PR(V82A) was able to compensate for the loss of interaction with inhibitor caused by mutation, in agreement with kinetic data, but substrate analogs have more flexibility than the drugs to accommodate the structural changes caused by mutation. Hence, these structures help to explain how HIV can develop drug resistance while retaining the ability of PR to hydrolyze natural substrates.
Figures



Similar articles
-
Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.Proteins. 2007 Apr 1;67(1):232-42. doi: 10.1002/prot.21304. Proteins. 2007. PMID: 17243183
-
High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains.J Mol Biol. 2004 Apr 23;338(2):341-52. doi: 10.1016/j.jmb.2004.02.052. J Mol Biol. 2004. PMID: 15066436
-
Crystal structures of HIV protease V82A and L90M mutants reveal changes in the indinavir-binding site.Eur J Biochem. 2004 Apr;271(8):1516-24. doi: 10.1111/j.1432-1033.2004.04060.x. Eur J Biochem. 2004. PMID: 15066177
-
Beyond darunavir: recent development of next generation HIV-1 protease inhibitors to combat drug resistance.Chem Commun (Camb). 2022 Oct 20;58(84):11762-11782. doi: 10.1039/d2cc04541a. Chem Commun (Camb). 2022. PMID: 36200462 Free PMC article. Review.
-
HIV protease: enzyme function and drug resistance.Vitam Horm. 2000;58:213-56. doi: 10.1016/s0083-6729(00)58026-1. Vitam Horm. 2000. PMID: 10668400 Review.
Cited by
-
Identification of folding preferences of cleavage junctions of HIV-1 precursor proteins for regulation of cleavability.J Mol Model. 2011 Feb;17(2):391-9. doi: 10.1007/s00894-010-0739-z. Epub 2010 May 18. J Mol Model. 2011. PMID: 20480379
-
Structural basis for a hand-like site in the calcium sensor CatchER with fast kinetics.Acta Crystallogr D Biol Crystallogr. 2013 Dec;69(Pt 12):2309-19. doi: 10.1107/S0907444913021306. Epub 2013 Nov 19. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24311573 Free PMC article.
-
Nine crystal structures determine the substrate envelope of the MDR HIV-1 protease.Protein J. 2011 Mar;30(3):173-83. doi: 10.1007/s10930-011-9316-2. Protein J. 2011. PMID: 21394574
-
Structural Insights to Human Immunodeficiency Virus (HIV-1) Targets and Their Inhibition.Adv Exp Med Biol. 2021;1322:63-95. doi: 10.1007/978-981-16-0267-2_3. Adv Exp Med Biol. 2021. PMID: 34258737
-
Comprehending the Structure, Dynamics, and Mechanism of Action of Drug-Resistant HIV Protease.ACS Omega. 2023 Mar 7;8(11):9748-9763. doi: 10.1021/acsomega.2c08279. eCollection 2023 Mar 21. ACS Omega. 2023. PMID: 36969469 Free PMC article. Review.
References
-
- Darke PL, Nutt RF, Brady SF, Garsky VM, Ciccarone TM, Leu CT, et al. HIV-1 protease specificity of peptide cleavage is sufficient for processing of Gag and Pol polyproteins. Biochem Biophys Res Commun. 1988;156:297–303. - PubMed
-
- Oroszlan S, Luftig RB. Retroviral proteases. Curr Top Microbiol Immunol. 1990;157:153–185. - PubMed
-
- Wlodawer A, Erickson JW. Structure-based inhibitors of HIV-1 protease. Annu Rev Biochem. 1993;62:543–585. - PubMed
-
- Erickson JW, Gulnik SV, Markowitz M. Protease inhibitors: resistance, cross-resistance, fitness and the choice of initial and salvage therapies. AIDS Res Hum Retroviruses. 1999;13:S189–S204. - PubMed
-
- Shafer RW, Winters MA, Palmer S, Merigan TC. Multiple concurrent reverse transcriptase and protease mutations and multidrug resistance of HIV-1 isolates from heavily treated patients. Ann Intern Med. 1998;128:906–911. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

