Antibody vs. HIV in a clash of evolutionary titans
- PMID: 16219699
- PMCID: PMC1257708
- DOI: 10.1073/pnas.0505126102
Antibody vs. HIV in a clash of evolutionary titans
Abstract
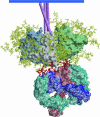
HIV has evolved many strategies to avoid neutralizing antibody responses, particularly to conserved regions on the external glycoprotein spikes of the virus. Nevertheless, a small number of antibodies have been evolved by the human immune system to recognize conserved parts of the glycoproteins, and therefore, have broadly neutralizing cross-strain activities. These antibodies constitute important tools in the quest to design immunogens that can elicit broadly neutralizing antibodies in humans and hence contribute to an effective HIV vaccine. Crystallographic analyses of the antibodies, in many cases in an antigen-complexed form, have revealed novel and, in some instances, remarkable structural adaptations to attain virus recognition. Antibodies, like HIV, can evolve relatively rapidly through mutation and selection. It seems that the structures of these broadly neutralizing antibodies bear witness to a heroic struggle between two titans of rapid evolution.
Figures

References
-
- Korber, B., Muldoon, M., Theiler, J., Gao, F., Gupta, R., Lapedes, A., Hahn, B. H., Wolinsky, S. & Bhattacharya, T. (2000) Science 288, 1789–1796. - PubMed
-
- AIDS Epidemic Update, December 2004 (Joint United Nations Programme on HIV/AIDS, Geneva), www.unaids.org/wad2004/report.html. - PubMed
-
- Wyatt, R. & Sodroski, J. (1998) Science 280, 1884–1888. - PubMed
-
- Evans, D. T. & Desrosiers, R. C. (2001) Immunol. Rev. 183, 141–158. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

