Nuclear-mitochondrial epistasis and drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds
- PMID: 16219776
- PMCID: PMC1456161
- DOI: 10.1534/genetics.105.046698
Nuclear-mitochondrial epistasis and drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds
Abstract
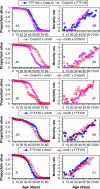
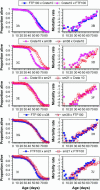
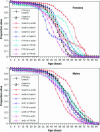
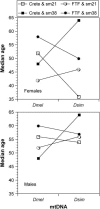
Under the mitochondrial theory of aging, physiological decline with age results from the accumulated cellular damage produced by reactive oxygen species generated during electron transport in the mitochondrion. A large body of literature has documented age-specific declines in mitochondrial function that are consistent with this theory, but relatively few studies have been able to distinguish cause from consequence in the association between mitochondrial function and aging. Since mitochondrial function is jointly encoded by mitochondrial (mtDNA) and nuclear genes, the mitochondrial genetics of aging should be controlled by variation in (1) mtDNA, (2) nuclear genes, or (3) nuclear-mtDNA interactions. The goal of this study was to assess the relative contributions of these factors in causing variation in Drosophila longevity. We compared strains of flies carrying mtDNAs with varying levels of divergence: two strains from Zimbabwe (<20 bp substitutions between mtDNAs), strains from Crete and the United States (approximately 20-40 bp substitutions between mtDNAs), and introgression strains of Drosophila melanogaster carrying mtDNA from Drosophila simulans in a D. melanogaster Oregon-R chromosomal background (>500 silent and 80 amino acid substitutions between these mtDNAs). Longevity was studied in reciprocal cross genotypes between pairs of these strains to test for cytoplasmic (mtDNA) factors affecting aging. The intrapopulation crosses between Zimbabwe strains show no difference in longevity between mtDNAs; the interpopulation crosses between Crete and the United States show subtle but significant differences in longevity; and the interspecific introgression lines showed very significant differences between mtDNAs. However, the genotypes carrying the D. simulans mtDNA were not consistently short-lived, as might be predicted from the disruption of nuclear-mitochondrial coadaptation. Rather, the interspecific mtDNA strains showed a wide range of variation that flanked the longevities seen between intraspecific mtDNAs, resulting in very significant nuclear x mtDNA epistatic interaction effects. These results suggest that even "defective" mtDNA haplotypes could extend longevity in different nuclear allelic backgrounds, which could account for the variable effects attributable to mtDNA haplogroups in human aging.
Figures

Similar articles
-
G×G×E for lifespan in Drosophila: mitochondrial, nuclear, and dietary interactions that modify longevity.PLoS Genet. 2014 May 15;10(5):e1004354. doi: 10.1371/journal.pgen.1004354. eCollection 2014. PLoS Genet. 2014. PMID: 24832080 Free PMC article.
-
Mitochondrial-nuclear epistasis: implications for human aging and longevity.Ageing Res Rev. 2011 Apr;10(2):238-52. doi: 10.1016/j.arr.2010.06.003. Epub 2010 Jun 25. Ageing Res Rev. 2011. PMID: 20601194 Free PMC article. Review.
-
Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila.Evolution. 2010 Dec;64(12):3364-79. doi: 10.1111/j.1558-5646.2010.01077.x. Evolution. 2010. PMID: 20624176 Free PMC article.
-
Mitochondrial-Nuclear Interactions Mediate Sex-Specific Transcriptional Profiles in Drosophila.Genetics. 2016 Oct;204(2):613-630. doi: 10.1534/genetics.116.192328. Epub 2016 Aug 24. Genetics. 2016. PMID: 27558138 Free PMC article.
-
Drosophila simulans as a novel model for studying mitochondrial metabolism and aging.Exp Gerontol. 2005 Oct;40(10):763-73. doi: 10.1016/j.exger.2005.07.014. Exp Gerontol. 2005. PMID: 16169180 Review.
Cited by
-
Ninety years of Drosophila melanogaster hybrids.Genetics. 2010 Sep;186(1):1-8. doi: 10.1534/genetics.110.121459. Genetics. 2010. PMID: 20855573 Free PMC article.
-
Maize cytolines as models to study the impact of different cytoplasms on gene expression under heat stress conditions.BMC Plant Biol. 2023 Jan 2;23(1):4. doi: 10.1186/s12870-022-04023-8. BMC Plant Biol. 2023. PMID: 36588161 Free PMC article.
-
New Insights into Mitochondrial-Nuclear Interactions Revealed through Analysis of Small RNAs.Genome Biol Evol. 2022 Feb 4;14(2):evac023. doi: 10.1093/gbe/evac023. Genome Biol Evol. 2022. PMID: 35143645 Free PMC article.
-
G × G × E effect on phenotype expression in a non-conventional model organism, the unicellular slime mould Physarum polycephalum.Biol Lett. 2023 Feb;19(2):20220494. doi: 10.1098/rsbl.2022.0494. Epub 2023 Feb 15. Biol Lett. 2023. PMID: 36789533 Free PMC article.
-
Mitochondrial Recombination Reveals Mito-Mito Epistasis in Yeast.Genetics. 2018 May;209(1):307-319. doi: 10.1534/genetics.117.300660. Epub 2018 Mar 12. Genetics. 2018. PMID: 29531011 Free PMC article.
References
-
- Agarwal, S., and R. S. Sohal, 1996. Relationship between susceptibility to protein oxidation, aging, and maximum life span potential of different species. Exp. Gerontol. 31: 365–372. - PubMed
-
- Balaban, R. S., S. Nemoto and T. Finkel, 2005. Mitochondria, oxidants, and aging. Cell 120: 483–495. - PubMed
-
- Ballard, J. W. O., 2000. Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J. Mol. Evol. 51: 48–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

