Drosophila target of rapamycin kinase functions as a multimer
- PMID: 16219781
- PMCID: PMC1456163
- DOI: 10.1534/genetics.105.051979
Drosophila target of rapamycin kinase functions as a multimer
Abstract
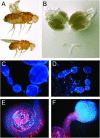
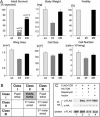
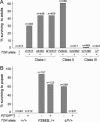
Target of rapamycin (TOR) is a conserved regulator of cell growth and metabolism that integrates energy, growth factor, and nutrient signals. The 280-kDa TOR protein functions as the catalytic component of two large multiprotein complexes and consists of an N-terminal HEAT-repeat domain and a C-terminal Ser/Thr kinase domain. Here we describe an allelic series of mutations in the Drosophila Tor gene and show that combinations of mutations in the HEAT and kinase domains of TOR display the rare genetic phenomenon of intragenic complementation, in which two or more defective proteins assemble to form a functional multimer. We present biochemical evidence that TOR self-associates in vivo and show that this multimerization is unaffected by positive or negative signals upstream of TOR. Consistent with multimerization of TOR, recessive mutations in the HEAT and kinase domains can dominantly interfere with wild-type TOR function in cells lacking TSC1 or TSC2. TOR multimerization thus partially accounts for the high apparent molecular weight of TOR complexes and offers novel therapeutic strategies for pathologies stemming from TOR hyperactivity.
Figures

Similar articles
-
Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling.Nat Cell Biol. 2002 Sep;4(9):699-704. doi: 10.1038/ncb847. Nat Cell Biol. 2002. PMID: 12172555
-
Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase.Proc Natl Acad Sci U S A. 2004 Sep 14;101(37):13489-94. doi: 10.1073/pnas.0405659101. Epub 2004 Sep 1. Proc Natl Acad Sci U S A. 2004. PMID: 15342917 Free PMC article.
-
The phosphatase subunit tap42 functions independently of target of rapamycin to regulate cell division and survival in Drosophila.Genetics. 2005 Jun;170(2):733-40. doi: 10.1534/genetics.104.039909. Epub 2005 Mar 31. Genetics. 2005. PMID: 15802506 Free PMC article.
-
TOR signaling: an odyssey from cellular stress to the cell growth machinery.Curr Biol. 2005 Feb 22;15(4):R139-41. doi: 10.1016/j.cub.2005.02.015. Curr Biol. 2005. PMID: 15723787 Review.
-
Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression.Oncogene. 2004 Apr 19;23(18):3151-71. doi: 10.1038/sj.onc.1207542. Oncogene. 2004. PMID: 15094765 Review.
Cited by
-
Spargel/dPGC-1 is essential for oogenesis and nutrient-mediated ovarian growth in Drosophila.Dev Biol. 2019 Oct 15;454(2):97-107. doi: 10.1016/j.ydbio.2019.06.020. Epub 2019 Jun 25. Dev Biol. 2019. PMID: 31251895 Free PMC article.
-
Spargel/dPGC-1 is a new downstream effector in the insulin-TOR signaling pathway in Drosophila.Genetics. 2013 Oct;195(2):433-41. doi: 10.1534/genetics.113.154583. Epub 2013 Aug 9. Genetics. 2013. PMID: 23934892 Free PMC article.
-
Regulation of TORC1 by Rag GTPases in nutrient response.Nat Cell Biol. 2008 Aug;10(8):935-45. doi: 10.1038/ncb1753. Epub 2008 Jul 6. Nat Cell Biol. 2008. PMID: 18604198 Free PMC article.
-
Specific roles of Target of rapamycin in the control of stem cells and their progeny in the Drosophila ovary.Development. 2010 Jul;137(13):2117-26. doi: 10.1242/dev.050351. Epub 2010 May 26. Development. 2010. PMID: 20504961 Free PMC article.
-
Integrated stress response signaling acts as a metabolic sensor in fat tissues to regulate oocyte maturation and ovulation.Cell Rep. 2024 Mar 26;43(3):113863. doi: 10.1016/j.celrep.2024.113863. Epub 2024 Mar 7. Cell Rep. 2024. PMID: 38457339 Free PMC article.
References
-
- Andrade, M. A., and P. Bork, 1995. HEAT repeats in the Huntington's disease protein. Nat. Genet. 11: 115–116. - PubMed
-
- Andrade, M. A., C. Petosa, S. I. O'Donoghue, C. W. Muller and P. Bork, 2001. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309: 1–18. - PubMed
-
- Bakkenist, C. J., and M. B. Kastan, 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

