Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development
- PMID: 16219919
- PMCID: PMC1255978
- DOI: 10.1104/pp.105.065748
Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development
Abstract
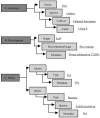
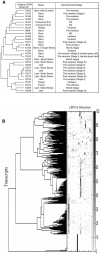
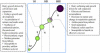
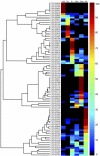
We report the analysis and annotation of 146,075 expressed sequence tags from Vitis species. The majority of these sequences were derived from different cultivars of Vitis vinifera, comprising an estimated 25,746 unique contig and singleton sequences that survey transcription in various tissues and developmental stages and during biotic and abiotic stress. Putatively homologous proteins were identified for over 17,752 of the transcripts, with 1,962 transcripts further subdivided into one or more Gene Ontology categories. A simple structured vocabulary, with modules for plant genotype, plant development, and stress, was developed to describe the relationship between individual expressed sequence tags and cDNA libraries; the resulting vocabulary provides query terms to facilitate data mining within the context of a relational database. As a measure of the extent to which characterized metabolic pathways were encompassed by the data set, we searched for homologs of the enzymes leading from glycolysis, through the oxidative/nonoxidative pentose phosphate pathway, and into the general phenylpropanoid pathway. Homologs were identified for 65 of these 77 enzymes, with 86% of enzymatic steps represented by paralogous genes. Differentially expressed transcripts were identified by means of a stringent believability index cutoff of > or =98.4%. Correlation analysis and two-dimensional hierarchical clustering grouped these transcripts according to similarity of expression. In the broadest analysis, 665 differentially expressed transcripts were identified across 29 cDNA libraries, representing a range of developmental and stress conditions. The groupings revealed expected associations between plant developmental stages and tissue types, with the notable exception of abiotic stress treatments. A more focused analysis of flower and berry development identified 87 differentially expressed transcripts and provides the basis for a compendium that relates gene expression and annotation to previously characterized aspects of berry development and physiology. Comparison with published results for select genes, as well as correlation analysis between independent data sets, suggests that the inferred in silico patterns of expression are likely to be an accurate representation of transcript abundance for the conditions surveyed. Thus, the combined data set reveals the in silico expression patterns for hundreds of genes in V. vinifera, the majority of which have not been previously studied within this species.
Figures


References
-
- Ablett E, Seaton G, Scott K, Shelton D, Graham MW, Baverstock P, Lee LS, Henry R (2000) Analysis of grape ESTs: global gene expression patterns in leaf and berry. Plant Sci 159: 87–95 - PubMed
-
- Akashi K, Nishimura N, Ishida Y, Yokota A (2004) Potent hydroxy radical-scavenging activity of drought-induced type-2 metallothionein in wild watermelon. Biochem Biophys Res Commun 323: 72–78 - PubMed
-
- Aldini G, Carini M, Piccoli A, Rossoni G, Facino RM (2003) Procyanidins from grape seeds protect endothelial cells from peroxynitrite damage and enhance endothelium-dependent relaxation in human artery: new evidences for cardio-protection. Life Sci 73: 2883–2898 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

