GERMINATE. a generic database for integrating genotypic and phenotypic information for plant genetic resource collections
- PMID: 16219922
- PMCID: PMC1255981
- DOI: 10.1104/pp.105.065201
GERMINATE. a generic database for integrating genotypic and phenotypic information for plant genetic resource collections
Abstract
The extensive germplasm resource collections that are now available for major crop plants and their wild relatives will increasingly provide valuable biological and bioinformatics resources for plant physiologists and geneticists to dissect the molecular basis of key traits and to develop highly adapted plant material to sustain future breeding programs. A key to the efficient deployment of these resources is the development of information systems that will enable the collection and storage of biological information for these plant lines to be integrated with the molecular information that is now becoming available through the use of high-throughput genomics and post-genomics technologies. The GERMINATE database has been designed to hold a diverse variety of data types, ranging from molecular to phenotypic, and to allow querying between such data for any plant species. Data are stored in GERMINATE in a technology-independent manner, such that new technologies can be accommodated in the database as they emerge, without modification of the underlying schema. Users can access data in GERMINATE databases either via a lightweight Perl-CGI Web interface or by the more complex Genomic Diversity and Phenotype Connection software. GERMINATE is released under the GNU General Public License and is available at http://germinate.scri.sari.ac.uk/germinate/.
Figures
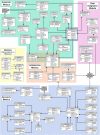

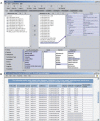
References
-
- Aharoni A, Vorst O (2002) DNA microarrays for functional genomics. Plant Mol Biol 48: 99–118 - PubMed
-
- Alercia A, Diulgheroff S, Metz T (2001) FAO/IPGRI Multi-Crop Passport Descriptors. http://www.ipgri.cgiar.org/publications (June 26, 2003)
-
- Brettschneider R (1998) RFLP analysis. In A Karp, PG Isaac, DS Ingram, eds, Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp 83–95
-
- Bruskiewich RM, Cosico AB, Eusebio W, Portugal AM, Ramos LM, Reyes MT, Sallan MA, Ulat VJ, Wang X, McNally KL, et al (2003) Linking genotype to phenotype: the International Rice Information System (IRIS). Bioinformatics (Suppl 1) 19: i63–i65 - PubMed
-
- Casstevens TM, Buckler ES (2004) GDPC: connecting researchers with multiple integrated data sources. Bioinformatics 20: 2839–2840 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

