Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice
- PMID: 16220146
- PMCID: PMC1250929
- DOI: 10.1371/journal.ppat.0010011
Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice
Abstract
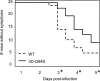
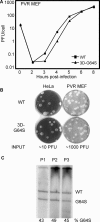
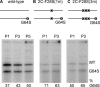
RNA viruses have high error rates, and the resulting quasispecies may aid survival of the virus population in the presence of selective pressure. Therefore, it has been theorized that RNA viruses require high error rates for survival, and that a virus with high fidelity would be less able to cope in complex environments. We previously isolated and characterized poliovirus with a mutation in the viral polymerase, 3D-G64S, which confers resistance to mutagenic nucleotide analogs via increased fidelity. The 3D-G64S virus was less pathogenic than wild-type virus in poliovirus-receptor transgenic mice, even though only slight growth defects were observed in tissue culture. To determine whether the high-fidelity phenotype of the 3D-G64S virus could decrease its fitness under a defined selective pressure, we compared growth of the 3D-G64S virus and 3D wild-type virus in the context of a revertible attenuating point mutation, 2C-F28S. Even with a 10-fold input advantage, the 3D-G64S virus was unable to compete with 3D wild-type virus in the context of the revertible attenuating mutation; however, in the context of a non-revertible version of the 2C-F28S attenuating mutation, 3D-G64S virus matched the replication of 3D wild-type virus. Therefore, the 3D-G64S high-fidelity phenotype reduced viral fitness under a defined selective pressure, making it likely that the reduced spread in murine tissue could be caused by the increased fidelity of the viral polymerase.
Conflict of interest statement
Figures


References
-
- Domingo E, Holland JJ. A virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. - PubMed
-
- Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. - PubMed
-
- Domingo E, Menendez-Arias L, Holland JJ. RNA virus fitness. Rev Med Virol. 1997;7:87–96. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

