Nonnatural amino acid incorporation into the methionine 214 position of the metzincin Pseudomonas aeruginosa alkaline protease
- PMID: 16221305
- PMCID: PMC1266349
- DOI: 10.1186/1471-2091-6-21
Nonnatural amino acid incorporation into the methionine 214 position of the metzincin Pseudomonas aeruginosa alkaline protease
Abstract
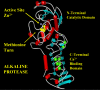
Background: The alkaline protease from Pseudomonas aeruginosa (AprA) is a member of the metzincin superfamily of metalloendoproteases. A key feature of these proteases is a conserved methionine-containing 1,4-tight beta turn at the base of the active site zinc binding region.
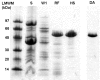
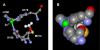
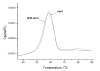
Results: To explore the invariant methionine position in this class of protease, incorporation of a nonnatural fluorinated methionine, L-difluoromethionine (DFM), into this site was accomplished. Although overproduction of the N-terminal catalytic fragment of AprA resulted in protein aggregates which could not be resolved, successful heterologous production of the entire AprA was accomplished in the presence and absence of the nonnatural amino acid. DFM incorporation was found to only slightly alter the enzyme kinetics of AprA. In addition, differential scanning calorimetry indicated no significant alteration in the thermal stability of the modified enzyme.
Conclusion: Although invariant in all metzincin proteases, the methionine 214 position in AprA can be successfully replaced by the nonnatural amino acid DFM resulting in little effect on protein structure and function. This study indicates that the increased size of the methyl group by the introduction of two fluorines is still sufficiently non-sterically demanding, and bodes well for the application of DFM to biophysical studies of protein structure and function in this class of protease.
Figures


Similar articles
-
Inhibition of Pseudomonas aeruginosa virulence: characterization of the AprA-AprI interface and species selectivity.J Mol Biol. 2012 Jan 20;415(3):573-83. doi: 10.1016/j.jmb.2011.11.039. Epub 2011 Nov 29. J Mol Biol. 2012. PMID: 22154939
-
Alkaline protease contributes to pyocyanin production in Pseudomonas aeruginosa.FEMS Microbiol Lett. 2017 Apr 1;364(7). doi: 10.1093/femsle/fnx051. FEMS Microbiol Lett. 2017. PMID: 28333255
-
Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site.Biol Chem. 2009 Sep;390(9):875-81. doi: 10.1515/BC.2009.100. Biol Chem. 2009. PMID: 19558324
-
[Mechanism of action of proteinases of Pseudomonas aeruginosa].Pathol Biol (Paris). 1990 Dec;38(10):968-74. Pathol Biol (Paris). 1990. PMID: 2127091 Review. French.
-
The metzincins--topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases.Protein Sci. 1995 May;4(5):823-40. doi: 10.1002/pro.5560040502. Protein Sci. 1995. PMID: 7663339 Free PMC article. Review.
Cited by
-
Pseudomonas evades immune recognition of flagellin in both mammals and plants.PLoS Pathog. 2011 Aug;7(8):e1002206. doi: 10.1371/journal.ppat.1002206. Epub 2011 Aug 25. PLoS Pathog. 2011. PMID: 21901099 Free PMC article.
-
Non-Canonical Amino Acids in Analyses of Protease Structure and Function.Int J Mol Sci. 2023 Sep 13;24(18):14035. doi: 10.3390/ijms241814035. Int J Mol Sci. 2023. PMID: 37762340 Free PMC article. Review.
-
Residue-Specific Incorporation of Noncanonical Amino Acids in Auxotrophic Hosts: Quo Vadis?.Chem Rev. 2025 May 28;125(10):4840-4932. doi: 10.1021/acs.chemrev.4c00280. Epub 2025 May 16. Chem Rev. 2025. PMID: 40378355 Free PMC article. Review.
-
On the relevance of the Met-turn methionine in metzincins.J Biol Chem. 2010 Apr 30;285(18):13951-7. doi: 10.1074/jbc.M109.083378. Epub 2010 Mar 4. J Biol Chem. 2010. PMID: 20202937 Free PMC article.
References
-
- Bode W, Gomis-Ruth FX, Stocker W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the 'metzincins'. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-I. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

