Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea
- PMID: 16221764
- PMCID: PMC1266117
- DOI: 10.1073/pnas.0506750102
Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea
Abstract
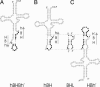
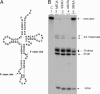
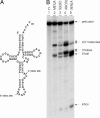
Members of the three kingdoms of life contain tRNA genes with introns. The introns in pre-tRNAs of Bacteria are self-splicing, whereas introns in archaeal and eukaryal pre-tRNAs are removed by splicing endonucleases. We have studied the structures of the endonucleases of Archaea and the architecture of the sites recognized in their pre-tRNA substrates. Three endonuclease structures are known in the Archaea: a homotetramer in some Euryarchaea, a homodimer in other Euryarchaea, and a heterotetramer in the Crenarchaeota. The homotetramer cleaves only the canonical bulge-helix-bulge structure in its substrates. Variants of the substrate structure, termed bulge-helix-loops, appear in the pre-tRNAs of the Crenarcheota and Nanoarcheota. These variant structures can be cleaved only by the homodimer or heterotetramer forms of the endonucleases. Thus, the structures of the endonucleases and their substrates appear to have evolved together.
Figures



References
-
- Kjems, J., Jensen, J., Olesen, T. & Garrett, R. A. (1989) Can J. Microbiol. 35, 210-214. - PubMed
-
- Kjems, J. & Garrett, R. A. (1988) Cell 54, 693-703. - PubMed
-
- Fabbri, S., Fruscoloni, P., Bufardeci, E., Di Nicola Negri, E., Baldi, M. I., Attardi, D. G., Mattoccia, E. & Tocchini-Valentini, G. P. (1998) Science 280, 284-286. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

