Q/R site editing controls kainate receptor inhibition by membrane fatty acids
- PMID: 16221857
- PMCID: PMC6725705
- DOI: 10.1523/JNEUROSCI.2826-05.2005
Q/R site editing controls kainate receptor inhibition by membrane fatty acids
Abstract
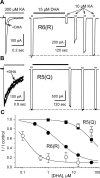
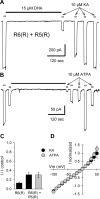
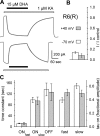
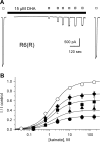
RNA editing within the pore loop controls the pharmacology and permeation properties of ion channels formed by neuronal AMPA and kainate receptor subunits. Genomic sequences for the glutamate receptor 2 (GluR2) subunit of AMPA receptors and the GluR5 and GluR6 subunits of kainate receptors all encode a neutral glutamine (Q) residue within the channel pore that can be converted by RNA editing to a positively charged arginine (R). Receptors comprised of unedited subunits are permeable to calcium and display inwardly rectifying current-voltage relationships, because of blocking of outward current by intracellular polyamines. In contrast, receptors that include edited subunits conduct less calcium, resist polyamine block, and have relatively linear current-voltage relationships. We showed previously that cis-unsaturated fatty acids, including arachidonic acid and docosahexanoic acid, exert a potent block of native kainate receptors as well as homomeric recombinant receptors formed by transfection of heterologous cells with cDNA for the GluR6(R) subunit. Here, we show that fatty acid blockade of recombinant homomeric and heteromeric kainate receptors is strongly dependent on editing at the Q/R site. Recombinant channels that include unedited subunits exhibit significantly weaker block than channels made up of fully edited subunits. Inhibition of fully edited channels is equivalent at voltages from -70 to +40 mV and is noncompetitive, consistent with allosteric regulation of channel function.
Figures



References
-
- Bernard A, Khrestchatisky M (1994) Assessing the extent of RNA editing in the TMII regions of GluR5 and GluR6 kainate receptors during rat brain development. J Neurochem 62: 2057-2060. - PubMed
-
- Bettler B, Boulter J, Hermans-Borgmeyer I, O'Shea-Greenfield A, Deneris E, Moll C, Borgmeyer U, Hollmann M, Heinemann S (1990) Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron 5: 583-595. - PubMed
-
- Bortolotto ZA, Clarke VR, Delany CM, Parry MC, Smolders I, Vignes M, Ho KH, Miu P, Brinton BT, Fantaske R, Ogden A, Gates M, Ornstein PL, Lodge D, Bleakman D, Collingridge GL (1999) Kainate receptors are involved in synaptic plasticity. Nature 402: 297-301. - PubMed
-
- Bowie D, Mayer ML (1995) Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron 15: 453-462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
