The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits
- PMID: 16221974
- PMCID: PMC1253832
- DOI: 10.1093/nar/gki887
The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits
Abstract
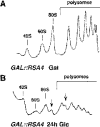
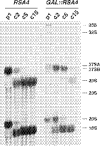
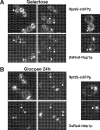
We report the characterization of a novel factor, Rsa4p (Ycr072cp), which is essential for the synthesis of 60S ribosomal subunits. Rsa4p is a conserved WD-repeat protein that seems to localize in the nucleolus. In vivo depletion of Rsa4p results in a deficit of 60S ribosomal subunits and the appearance of half-mer polysomes. Northern hybridization and primer extension analyses of pre-rRNA and mature rRNAs show that depletion of Rsa4p leads to the accumulation of the 27S, 25.5S and 7S pre-rRNAs, resulting in a reduction of the mature 25S and 5.8S rRNAs. Pulse-chase analyses of pre-rRNA processing reveal that, at least, this is due to a strong delay in the maturation of 27S pre-rRNA intermediates to mature 25S rRNA. Furthermore, depletion of Rsa4p inhibited the release of the pre-60S ribosomal particles from the nucleolus to the nucleoplasm, as judged by the predominantly nucleolar accumulation of the large subunit Rpl25-eGFP reporter construct. We propose that Rsa4p associates early with pre-60S ribosomal particles and provides a platform of interaction for correct processing of rRNA precursors and nucleolar release of 60S ribosomal subunits.
Figures






References
-
- Olson M.O., Dundr M., Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. - PubMed
-
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. - PubMed
-
- de la Cruz J., Kressler D., Linder P. Ribosomal subunit assembly. In: Olson M.O.J., editor. Nucleolus. Georgetown: Kluwer academic. Landes Bioscience/eurekah.com; 2004. pp. 258–285.
-
- Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. - PubMed
-
- Raué H.A. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine. In: Olson M.O.J., editor. Nucleolus. Georgetown: Kluwer academic. Landes Biosciences/Eurekah.com; 2004. pp. 199–222.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

