Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats
- PMID: 16221978
- PMCID: PMC1253834
- DOI: 10.1093/nar/gki898
Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats
Abstract
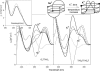
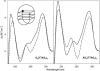
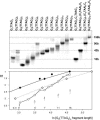
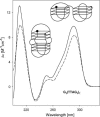
Secondary structures of the G-rich strand of human telomere DNA fragments G3(TTAG3)n, n = 1-16, have been studied by means of circular dichroism spectroscopy and PAGE, in solutions of physiological potassium cation concentrations. It has been found that folding of these fragments into tetraplexes as well as tetraplex thermostabilities and enthalpy values depend on the number of TTAG3 repeats. The suggested topologies include, e.g. antiparallel and parallel bimolecular tetraplexes, an intramolecular antiparallel tetraplex, a tetraplex consisting of three parallel chains and one antiparallel chain, a poorly stable parallel intramolecular tetraplex, and both parallel and antiparallel tetramolecular tetraplexes. G3(TTAG3)3 folds into a single, stable and very compact intramolecular antiparallel tetraplex. With an increasing repeat number, the fragment tetraplexes surprisingly are ever less thermostable and their migration and enthalpy decrease indicate increasing irregularities or domain splitting in their arrangements. Reduced stability and different topology of lengthy telomeric tails could contribute to the stepwise telomere shortening process.
Figures



References
-
- Blackburn E.H., Greider C.W. Telomeres. Plainview, NY, USA: Cold Spring Harbor Laboratory; 1995.
-
- Cech T.R., Nakamura T.M., Lingner J. Telomerase is a true reverse transcriptase. A review. Biochemistry (Mosc) 1997;62:1202–1205. - PubMed
-
- Henderson E., Hardin C.C., Walk S.K., Tinoco I.J., Blackburn E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine–guanine base pairs. Cell. 1987;51:899–908. - PubMed
-
- Choi K.-H., Choi B.-S. Formation of a hairpin structure by telomere 3′ overhang. Biochim. Biophys. Acta. 1994;1217:341–344. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

