Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development
- PMID: 16223478
- PMCID: PMC3869089
- DOI: 10.1016/j.ydbio.2005.09.024
Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development
Abstract
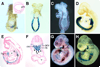
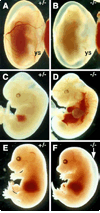
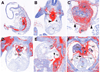
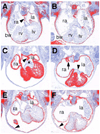
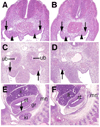
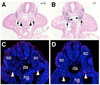
The Odd-skipped related 1 (Odd 1) gene encodes a zinc finger protein homologous to the Drosophila Odd-skipped class transcription factors that play critical roles in embryonic patterning and tissue morphogenesis. We have generated mice carrying a targeted null mutation in the Odd 1 gene and show that Odd 1 is essential for heart and intermediate mesoderm development. Odd 1(-/-) mutant mouse embryos fail to form atrial septum, display dilated atria with hypoplastic venous valves, and exhibit blood backflow from the heart into systemic veins. In contrast to other transcription factors implicated in atrial septum development, Odd 1 mRNA expression is restricted to the central dorsal domain of the atrial myocardium during normal heart development. Moreover, expression patterns of known key regulatory genes of atrial septum development, including Nkx2.5, Pitx2, and Tbx5, are unaltered in the developing heart in Odd 1(-/-) mutants compared to that of the wild-type littermates. Furthermore, homozygous Odd 1(-/-) mutant embryos exhibit complete agenesis of adrenal glands, metanephric kidneys, gonads, and defects in pericardium formation. Detailed molecular marker analyses show that key regulators of early intermediate mesoderm development, including Lhx1, Pax2, and Wt1, are all down-regulated and nephrogenic mesenchyme undergoes massive apoptosis, resulting in disruption of nephric duct elongation and failure of metanephric induction in the Odd 1(-/-) mutant embryos. These data provide new insights into the molecular mechanisms underlying heart morphogenesis and urogenital development.
Figures






References
-
- Anderson RH, Webb S, Brown NA. Clinical anatomy of the atrial septum with reference to its developmental components. Clin. Anat. 1999;12:362–374. - PubMed
-
- Armstrong JF, Pritchard-Jones K, Bickmore WA, Hastie ND, Bard JB. The expression of the wilms’ tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 1993;40:85–97. - PubMed
-
- Arrechedera H, Alvarez M, Strauss M, Ayesta C. Origin of mesenchymal tissue in the septum primum: a structural and ultrastructural study. J. Mol. Cell. Cardiol. 1987;19:641–651. - PubMed
-
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE. Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nature Genet. 1997;15:30–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

