A simple method for displaying recalcitrant proteins on the surface of bacteriophage lambda
- PMID: 16224099
- PMCID: PMC1258178
- DOI: 10.1093/nar/gni158
A simple method for displaying recalcitrant proteins on the surface of bacteriophage lambda
Abstract
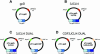
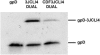
Bacteriophage lambda (lambda) permits the display of many foreign peptides and proteins on the gpD major coat protein. However, some recombinant derivatives of gpD are incompatible with the assembly of stable phage particles. This presents a limitation to current lambda display systems. Here we describe a novel, plasmid-based expression system in which gpD deficient lambda lysogens can be co-complemented with both wild-type and recombinant forms of gpD. This dual expression system permits the generation of mosaic phage particles that contain otherwise recalcitrant recombinant gpD fusion proteins. Overall, this improved gpD display system is expected to permit the expression of a wide variety of peptides and proteins on the surface of bacteriophage lambda and to facilitate the use of modified lambda phage vectors in mammalian gene transfer applications.
Figures


References
-
- Murialdo H., Ray P.N. Model for arrangement of minor structural proteins in head of bacteriophage lambda. Nature. 1975;257:815–817. - PubMed
-
- Sternberg N., Weisberg R. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J. Mol. Biol. 1977;117:733–759. - PubMed
-
- Yang F., Forrer P., Dauter Z., Conway J.F., Cheng N., Cerritelli M.E., Steven A.C., Pluckthun A., Wlodawer A. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat. Struct. Biol. 2000;7:230–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

