Method for multiplex cellular detection of mRNAs using quantum dot fluorescent in situ hybridization
- PMID: 16224100
- PMCID: PMC1258180
- DOI: 10.1093/nar/gni162
Method for multiplex cellular detection of mRNAs using quantum dot fluorescent in situ hybridization
Abstract
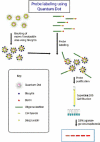
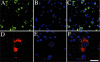
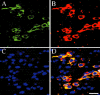
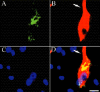
The photostability and narrow emission spectra of non-organic quantum dot fluorophores (QDs) make them desirable candidates for fluorescent in situ hybridization (FISH) to study the expression of specific mRNA transcripts. We developed a novel method for direct QD labeling of modified oligonucleotide probes through streptavidin and biotin interactions, as well as protocols for their use in multiple-label FISH. We validated this technique in mouse brainstem sections. The subcellular localization of the vesicular monoamine transporter (Vmat2) mRNA corresponds when using probes labeled with two different QDs in the same hybridization. We developed protocols for combined direct QD FISH and QD immunohistochemical labeling within the same neurons as well as for simultaneous study of the subcellular distribution of multiple mRNA targets. We demonstrated increased sensitivity of FISH using QDs in comparison with organic fluorophores. These techniques gave excellent histological results both for multiplex FISH and combined FISH and immunohistochemistry. This approach can facilitate the ultrasensitive simultaneous study of multiple mRNA and protein markers in tissue culture and histological section.
Figures




References
-
- Burgunder J.M., Young W.S., III The distribution of thalamic projection neurons containing cholecystokinin messenger RNA, using in situ hybridization histochemistry and retrograde labeling. Brain Res. 1988;464:179–189. - PubMed
-
- Durrant I., Brunning S., Eccleston L., Chadwick P., Cunningham M. Fluorescein as a label for non-radioactive in situ hybridization. Histochem. J. 1995;27:94–99. - PubMed
-
- Hermanson O., Ericson H., Sanchez-Watts G., Watts A.G., Blomqvist A. Autoradiographic visualization of 35S-labeled cRNA probes combined with immunoperoxidase detection of choleragenoid: a double-labeling light microscopic method for in situ hybridization and retrograde tract tracing. J. Histochem. Cytochem. 1994;42:827–831. - PubMed
-
- Chartrand P., Bertrand E., Singer R.H., Long R.M. Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 2000;318:493–506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

