Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis
- PMID: 16224102
- PMCID: PMC1258174
- DOI: 10.1093/nar/gki901
Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis
Abstract
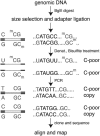
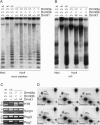
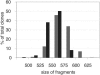
We describe a large-scale random approach termed reduced representation bisulfite sequencing (RRBS) for analyzing and comparing genomic methylation patterns. BglII restriction fragments were size-selected to 500-600 bp, equipped with adapters, treated with bisulfite, PCR amplified, cloned and sequenced. We constructed RRBS libraries from murine ES cells and from ES cells lacking DNA methyltransferases Dnmt3a and 3b and with knocked-down (kd) levels of Dnmt1 (Dnmt[1(kd),3a-/-,3b-/-]). Sequencing of 960 RRBS clones from Dnmt[1(kd),3a-/-,3b-/-] cells generated 343 kb of non-redundant bisulfite sequence covering 66212 cytosines in the genome. All but 38 cytosines had been converted to uracil indicating a conversion rate of >99.9%. Of the remaining cytosines 35 were found in CpG and 3 in CpT dinucleotides. Non-CpG methylation was >250-fold reduced compared with wild-type ES cells, consistent with a role for Dnmt3a and/or Dnmt3b in CpA and CpT methylation. Closer inspection revealed neither a consensus sequence around the methylated sites nor evidence for clustering of residual methylation in the genome. Our findings indicate random loss rather than specific maintenance of methylation in Dnmt[1(kd),3a-/-,3b-/-] cells. Near-complete bisulfite conversion and largely unbiased representation of RRBS libraries suggest that random shotgun bisulfite sequencing can be scaled to a genome-wide approach.
Figures

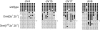
References
-
- Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. - PubMed
-
- Robertson K.D., Wolffe A.P. DNA methylation in health and disease. Nature Rev. Genet. 2000;1:11–19. - PubMed
-
- Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet. 1997;13:323–329. - PubMed
-
- Bestor T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. - PubMed
-
- Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet. 2003;33(Suppl):245–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

