Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein
- PMID: 16227265
- PMCID: PMC1262560
- DOI: 10.1128/JVI.79.21.13442-13453.2005
Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein
Abstract
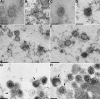
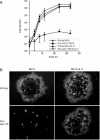
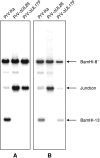
Homologs of the UL17 gene of the alphaherpesvirus herpes simplex virus 1 (HSV-1) are conserved in all three subfamilies of herpesviruses. However, only the HSV-1 protein has so far been characterized in any detail. To analyze UL17 of pseudorabies virus (PrV) the complete 597-amino-acid protein was expressed in Escherichia coli and used for rabbit immunization. The antiserum recognized a 64-kDa protein in PrV-infected cell lysates and purified virions, identifying PrV UL17 as a structural virion component. In indirect immunofluorescence analyses of PrV-infected cells the protein was predominantly found in the nucleus. In electron microscopic studies after immunogold labeling of negatively stained purified virion preparations, UL17-specific label was detected on single, mostly damaged capsids, whereas complete virions and the majority of capsids were free of label. In ultrathin sections of infected cells, label was primarily found dispersed around scaffold-containing B-capsids, whereas on DNA-filled C-capsids it was located in the center. Empty intranuclear A-capsids were free of label, as were extracellular capsid-less L-particles. Functional characterization of PrV-DeltaUL17F, a deletion mutant lacking codons 23 to 444, demonstrated that cleavage of viral DNA into unit-length genomes was inhibited in the absence of UL17. In electron microscopic analyses of PrV-DeltaUL17F-infected RK13 cells, DNA-containing capsids were not detected, while numerous capsidless L-particles were observed. In summary, our data indicate that the PrV UL17 protein is an internal nucleocapsid protein necessary for DNA cleavage and packaging but suggest that the protein is not a prominent part of the tegument.
Figures





Similar articles
-
The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids.J Virol. 2006 Jul;80(13):6235-46. doi: 10.1128/JVI.02662-05. J Virol. 2006. PMID: 16775311 Free PMC article.
-
The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged.Virology. 1998 Dec 5;252(1):115-25. doi: 10.1006/viro.1998.9439. Virology. 1998. PMID: 9875322
-
Characterization of pseudorabies virus (PrV) cleavage-encapsidation proteins and functional complementation of PrV pUL32 by the homologous protein of herpes simplex virus type 1.J Virol. 2009 Apr;83(8):3930-43. doi: 10.1128/JVI.02636-08. Epub 2009 Feb 4. J Virol. 2009. PMID: 19193798 Free PMC article.
-
Transport and egress of herpes simplex virus in neurons.Rev Med Virol. 2008 Jan-Feb;18(1):35-51. doi: 10.1002/rmv.560. Rev Med Virol. 2008. PMID: 17992661 Review.
-
Herpesvirus Capsid Assembly and DNA Packaging.Adv Anat Embryol Cell Biol. 2017;223:119-142. doi: 10.1007/978-3-319-53168-7_6. Adv Anat Embryol Cell Biol. 2017. PMID: 28528442 Free PMC article. Review.
Cited by
-
Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids.Proc Natl Acad Sci U S A. 2009 Jun 16;106(24):9673-8. doi: 10.1073/pnas.0901514106. Epub 2009 Jun 1. Proc Natl Acad Sci U S A. 2009. PMID: 19487681 Free PMC article.
-
Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid.J Virol. 2006 Nov;80(21):10894-9. doi: 10.1128/JVI.01364-06. Epub 2006 Aug 18. J Virol. 2006. PMID: 16920825 Free PMC article.
-
Human Cytomegalovirus Egress: Overcoming Barriers and Co-Opting Cellular Functions.Viruses. 2021 Dec 22;14(1):15. doi: 10.3390/v14010015. Viruses. 2021. PMID: 35062219 Free PMC article. Review.
-
Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice.J Virol. 2006 Jun;80(11):5571-6. doi: 10.1128/JVI.02589-05. J Virol. 2006. PMID: 16699038 Free PMC article.
-
The axonal sorting activity of pseudorabies virus Us9 protein depends on the state of neuronal maturation.PLoS Pathog. 2020 Dec 28;16(12):e1008861. doi: 10.1371/journal.ppat.1008861. eCollection 2020 Dec. PLoS Pathog. 2020. PMID: 33370419 Free PMC article.
References
-
- Al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180:380-388. - PubMed
-
- Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 310:207-211. - PubMed
-
- Baines, J. D., and S. K. Weller. 2005. Cleavage and packaging of herpes simplex virus 1 DNA. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structures, and mechanism. Landes Biosciences, Georgetown, Tex.
-
- Beard, P. M., and J. D. Baines. 2004. The DNA cleavage and packaging protein encoded by the UL33 gene of herpes simplex virus 1 associates with capsids. Virology 324:475-482. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

