Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo
- PMID: 16227570
- PMCID: PMC1265836
- DOI: 10.1128/MCB.25.21.9165-9174.2005
Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo
Abstract
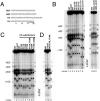
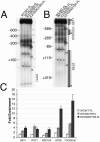
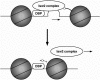
We have previously shown that Saccharomyces cerevisiae Isw2 complex slides nucleosomes to remodel chromatin in vivo. Our data suggested a model in which Isw2 complex binds the histone octamer and DNA separately to generate the force necessary for nucleosome movement. Here we find that the histone H4 "basic patch" is the only portion of any amino-terminal histone tail required for both target-specific association of Isw2 complex with chromatin and chromatin remodeling in vivo, whereas it is dispensable for basal levels of chromatin binding. Similarly, we find that nonremodeled chromatin structure and integrity of Isw2 complex are required only for target-specific association of Isw2 with chromatin. These data demonstrate fundamental differences between the target-specific and basal modes of chromatin binding by Isw2 complex in vivo and suggest that only the former involves contributions from DNA, histone H4, and sequence-specific DNA binding proteins. We propose a model for target recognition and chromatin remodeling by Isw2 complex in vivo.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
