Direct regulation of rRNA transcription by fibroblast growth factor 2
- PMID: 16227592
- PMCID: PMC1265826
- DOI: 10.1128/MCB.25.21.9419-9426.2005
Direct regulation of rRNA transcription by fibroblast growth factor 2
Abstract
Fibroblast growth factor 2 (FGF-2), which is highly expressed in developing tissues and malignant cells, regulates cell growth, differentiation, and migration. Five isoforms (18 to approximately 34 kDa) of FGF-2 are derived from alternative initiation codons of a single mRNA. The 18-kDa FGF-2 isoform is released from cells by a nonclassical secretory pathway and regulates gene expression by binding to cell surface receptors. This isoform also localizes to the nucleolus, raising the possibility that it may directly regulate ribosome biogenesis, a rate-limiting process in cell growth. Although several growth factors have been shown to accumulate in the nucleolus, their function and mechanism of action remain unclear. Here we show that 18-kDa FGF-2 interacts with upstream binding factor (UBF), an architectural transcription factor essential for rRNA transcription. The maximal activation of rRNA transcription in vitro by 18-kDa FGF-2 requires UBF. The 18-kDa FGF-2 localizes to rRNA genes and is necessary for the full activation of pre-rRNA synthesis in vivo. Our results demonstrate that 18-kDa FGF-2 directly regulates rRNA transcription.
Figures
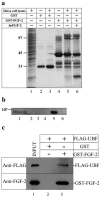

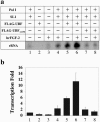
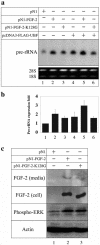

Similar articles
-
MXD1 localizes in the nucleolus, binds UBF and impairs rRNA synthesis.Oncotarget. 2016 Oct 25;7(43):69536-69548. doi: 10.18632/oncotarget.11766. Oncotarget. 2016. PMID: 27588501 Free PMC article.
-
RINT-1 interacts with MSP58 within nucleoli and plays a role in ribosomal gene transcription.Biochem Biophys Res Commun. 2016 Sep 16;478(2):873-80. doi: 10.1016/j.bbrc.2016.08.044. Epub 2016 Aug 13. Biochem Biophys Res Commun. 2016. PMID: 27530925
-
The transcription factor EGR1 localizes to the nucleolus and is linked to suppression of ribosomal precursor synthesis.PLoS One. 2014 May 1;9(5):e96037. doi: 10.1371/journal.pone.0096037. eCollection 2014. PLoS One. 2014. PMID: 24787739 Free PMC article.
-
The high molecular weight isoforms of basic fibroblast growth factor (FGF-2): an insight into an intracrine mechanism.FEBS Lett. 2000 Feb 18;468(1):6-10. doi: 10.1016/s0014-5793(00)01189-3. FEBS Lett. 2000. PMID: 10683430 Review.
-
Regulation of ribosomal RNA gene expression in porcine oocytes.Anim Reprod Sci. 2004 Jul;82-83:605-16. doi: 10.1016/j.anireprosci.2004.04.023. Anim Reprod Sci. 2004. PMID: 15271483 Review.
Cited by
-
CK2-mediated stimulation of Pol I transcription by stabilization of UBF-SL1 interaction.Nucleic Acids Res. 2006;34(17):4752-66. doi: 10.1093/nar/gkl581. Epub 2006 Sep 13. Nucleic Acids Res. 2006. PMID: 16971462 Free PMC article.
-
Overexpression of fibroblast growth factor receptor 2 in bone marrow mesenchymal stem cells enhances osteogenesis and promotes critical cranial bone defect regeneration.Front Cell Dev Biol. 2023 May 17;11:1208239. doi: 10.3389/fcell.2023.1208239. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37266455 Free PMC article.
-
The antiproliferative effect of FGF2 in K-Ras-driven tumor cells involves modulation of rRNA and the nucleolus.J Cell Sci. 2023 Nov 15;136(22):jcs260989. doi: 10.1242/jcs.260989. Epub 2023 Nov 30. J Cell Sci. 2023. PMID: 37921359 Free PMC article.
-
Nuclear trafficking of secreted factors and cell-surface receptors: new pathways to regulate cell proliferation and differentiation, and involvement in cancers.Cell Commun Signal. 2006 Oct 18;4:7. doi: 10.1186/1478-811X-4-7. Cell Commun Signal. 2006. PMID: 17049074 Free PMC article.
-
Cells resist starvation through a nutrient stress splice switch.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf525. doi: 10.1093/nar/gkaf525. Nucleic Acids Res. 2025. PMID: 40550515 Free PMC article.
References
-
- Bailly, K., F. Soulet, D. Leroy, F. Amalric, and G. Bouche. 2000. Uncoupling of cell proliferation and differentiation activities of basic fibroblast growth factor. FASEB J. 14:333-344. - PubMed
-
- Bikfalvi, A., S. Klein, G. Pintucci, and D. B. Rifkin. 1997. Biological roles of fibroblast growth factor-2. Endocr. Rev. 18:26-45. - PubMed
-
- Bonnet, H., O. Filhol, I. Truchet, P. Brethenou, C. Cochet, F. Amalric, and G. Bouche. 1996. Fibroblast growth factor-2 binds to the regulatory beta subunit of CK2 and directly stimulates CK2 activity toward nucleolin. J. Biol. Chem. 271:24781-24787. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
