Impaired immune responses and prolonged allograft survival in Sly1 mutant mice
- PMID: 16227612
- PMCID: PMC1265838
- DOI: 10.1128/MCB.25.21.9646-9660.2005
Impaired immune responses and prolonged allograft survival in Sly1 mutant mice
Abstract
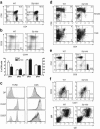
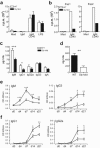
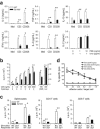
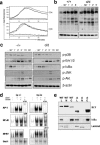
Adaptive immunity is crucial for protective host defense and the development of immunological disorders. SLY1 was recently identified as an X-chromosomal SH3 protein that is serine phosphorylated (Ser27) upon B-and T-cell receptor engagement. Here, we demonstrate that SLY1 is localized in the cytoplasm and the nucleus of immunocytes. We generated mice expressing a mutant version of SLY1 lacking Ser27 and a functional nuclear localization signal. The defective SLY1 (SLY1(d)) protein is localized exclusively in the cytoplasm. B- and T-cell proliferation is attenuated and T-cell cytokine production is severely reduced. Sly1(d/d) mice exhibit reduced lymphoid organ sizes, diminished marginal zone B-cell numbers, and severely impaired antibody responses against T-dependent and -independent antigens. Importantly, survival of semi-identical cardiac allografts was substantially prolonged in Sly1(d/d) mice. These results define SLY1 as an essential molecular component for the full activation of adaptive immunity.
Figures




References
-
- Astoul, E., A. D. Laurence, N. Totty, S. Beer, D. R. Alexander, and D. A. Cantrell. 2003. Approaches to define antigen receptor induced serine kinase signal transduction pathways. J. Biol. Chem. 278:9267-9275. - PubMed
-
- Beer, S., A. B. Simins, A. Schuster, and B. Holzmann. 2001. Molecular cloning and characterization of a novel SH3 protein (SLY) preferentially expressed in lymphoid cells. Biochim. Biophys. Acta 1520:89-93. - PubMed
-
- Cho, H. J., K. Takabayashi, P. M. Cheng, M. D. Nguyen, M. Corr, S. Tuck, and E. Raz. 2000. Immunostimulatory DNA-based vaccines induce cytotoxic lymphocyte activity by a T-helper cell-independent mechanism. Nat. Biotechnol. 18:509-514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
