FtsZ and the division of prokaryotic cells and organelles
- PMID: 16227976
- PMCID: PMC4757588
- DOI: 10.1038/nrm1745
FtsZ and the division of prokaryotic cells and organelles
Abstract
Binary fission of many prokaryotes as well as some eukaryotic organelles depends on the FtsZ protein, which self-assembles into a membrane-associated ring structure early in the division process. FtsZ is homologous to tubulin, the building block of the microtubule cytoskeleton in eukaryotes. Recent advances in genomics and cell-imaging techniques have paved the way for the remarkable progress in our understanding of fission in bacteria and organelles.
Conflict of interest statement
Competing interests statement
The author declares no competing financial interests.
Figures
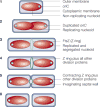
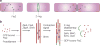



References
-
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. - PubMed
-
- Weiss DS. Bacterial cell division and the septal ring. Mol Microbiol. 2004;54:588–597. - PubMed
-
- Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol. 2004;58:19–29. - PubMed
-
- Sontag CA, Staley JT, Erickson HP. In vitro assembly and GTP hydrolysis by bacterial tubulins BtubA and BtubB. J Cell Biol. 2005;169:233–238. A member of the Chlamydia/Verrucomicrobia group of bacteria that lacks ftsZ contains instead two genes that are evolutionarily closer to tubulin than FtsZ. This paper investigates the assembly and nucleotide-binding properties of these two bacterial tubulins. - PMC - PubMed
-
- Glass JI, et al. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature. 2000;407:757–762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

