Probing the affinity of SecA for signal peptide in different environments
- PMID: 16229488
- PMCID: PMC3094106
- DOI: 10.1021/bi050882k
Probing the affinity of SecA for signal peptide in different environments
Abstract
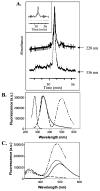
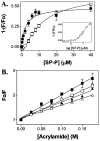
SecA, the peripheral subunit of the Escherichia coli preprotein translocase, interacts with a number of ligands during export, including signal peptides, membrane phospholipids, and nucleotides. Using fluorescence resonance energy transfer (FRET), we studied the interactions of wild-type (WT) and mutant SecAs with IAEDANS-labeled signal peptide, and how these interactions are modified in the presence of other transport ligands. We find that residues on the third alpha-helix in the preprotein cross-linking domain (PPXD) are important for the interaction of SecA and signal peptide. For SecA in aqueous solution, saturation binding data using FRET analysis fit a single-site binding model and yielded a Kd of 2.4 microM. FRET is inhibited for SecA in lipid vesicles relative to that in aqueous solution at a low signal peptide concentration. The sigmoidal nature of the binding curve suggests that SecA in lipids has two conformational states; our results do not support different oligomeric states of SecA. Using native gel electrophoresis, we establish signal peptide-induced SecA monomerization in both aqueous solution and lipid vesicles. Whereas the affinity of SecA for signal peptide in an aqueous environment is unaffected by temperature or the presence of nucleotides, in lipids the affinity decreases in the presence of ADP or AMP-PCP but increases at higher temperature. The latter finding is consistent with SecA existing in an elongated form while inserting the signal peptide into membranes.
Figures




Similar articles
-
Defining the solution state dimer structure of Escherichia coli SecA using Förster resonance energy transfer.Biochemistry. 2013 Apr 9;52(14):2388-401. doi: 10.1021/bi301217t. Epub 2013 Mar 29. Biochemistry. 2013. PMID: 23484952 Free PMC article.
-
Mapping of the signal peptide-binding domain of Escherichia coli SecA using Förster resonance energy transfer.Biochemistry. 2010 Feb 2;49(4):782-92. doi: 10.1021/bi901446r. Biochemistry. 2010. PMID: 20025247 Free PMC article.
-
Lipid and signal peptide-induced conformational changes within the C-domain of Escherichia coli SecA protein.Biochemistry. 2001 Feb 13;40(6):1835-43. doi: 10.1021/bi002058w. Biochemistry. 2001. PMID: 11327846
-
Oligomeric states of the SecA and SecYEG core components of the bacterial Sec translocon.Biochim Biophys Acta. 2007 Jan;1768(1):5-12. doi: 10.1016/j.bbamem.2006.08.013. Epub 2006 Aug 30. Biochim Biophys Acta. 2007. PMID: 17011510 Free PMC article. Review.
-
Structure and function of SecA, the preprotein translocase nanomotor.Biochim Biophys Acta. 2004 Nov 11;1694(1-3):67-80. doi: 10.1016/j.bbamcr.2004.06.003. Biochim Biophys Acta. 2004. PMID: 15546658 Review.
Cited by
-
Selective photoaffinity labeling identifies the signal peptide binding domain on SecA.J Mol Biol. 2007 Jan 19;365(3):637-48. doi: 10.1016/j.jmb.2006.10.027. Epub 2006 Nov 3. J Mol Biol. 2007. PMID: 17084862 Free PMC article.
-
Mapping of the SecA signal peptide binding site and dimeric interface by using the substituted cysteine accessibility method.J Bacteriol. 2013 Oct;195(20):4709-15. doi: 10.1128/JB.00661-13. Epub 2013 Aug 9. J Bacteriol. 2013. PMID: 23935053 Free PMC article.
-
Alignment of the protein substrate hairpin along the SecA two-helix finger primes protein transport in Escherichia coli.Proc Natl Acad Sci U S A. 2017 Aug 29;114(35):9343-9348. doi: 10.1073/pnas.1702201114. Epub 2017 Aug 10. Proc Natl Acad Sci U S A. 2017. PMID: 28798063 Free PMC article.
-
Characterization of the Escherichia coli SecA signal peptide-binding site.J Bacteriol. 2012 Jan;194(2):307-16. doi: 10.1128/JB.06150-11. Epub 2011 Nov 4. J Bacteriol. 2012. PMID: 22056930 Free PMC article.
-
In Vitro Interaction of the Housekeeping SecA1 with the Accessory SecA2 Protein of Mycobacterium tuberculosis.PLoS One. 2015 Jun 5;10(6):e0128788. doi: 10.1371/journal.pone.0128788. eCollection 2015. PLoS One. 2015. PMID: 26047312 Free PMC article.
References
-
- Kim J, Miller A, Wang L, Muller JP, Kendall DA. Evidence that SecB enhances the activity of SecA. Biochemistry. 2001;40:3674–80. - PubMed
-
- Driessen AJ, Fekkes P, van der Wolk JP. The Sec system. Curr Opin Microbiol. 1998;1:216–22. - PubMed
-
- Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

