Identification of a novel amidase motif in neutral ceramidase
- PMID: 16229686
- PMCID: PMC1360721
- DOI: 10.1042/BJ20050682
Identification of a novel amidase motif in neutral ceramidase
Abstract
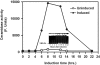
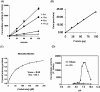
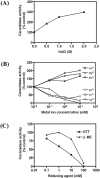
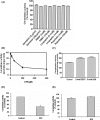
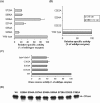
Neutral CDases (ceramidases) are newly identified enzymes with important roles in cell regulation, but little is known about their catalytic mechanisms. In the present study the full-length human neutral CDase was cloned and expressed in the yeast double-knockout strain Dypc1Dydc1, which lacks the yeast CDases YPC1p and YDC1p. Biochemical characterization of the human neutral CDase showed that the enzyme exhibited classical Michaelis-Menten kinetics, with an optimum activity at pH 7.5. Activity was enhanced by Na+ and Ca2+. Mg2+ and Mn2+ were somewhat stimulatory, but Zn2+, Cu2+ and Fe2+ inhibited the enzyme. Dithiothreitol and 2-mercaptoethanol dose-dependently inhibited neutral CDase. In order to identify which amino acids were involved in the catalytic action of neutral CDase, the purified enzyme was subjected to chemical modifications. It was observed that the serine residue modifier di-isopropyl fluorophosphate dose-dependently inhibited activity, implicating a serine residue in the catalytic action. From an alignment of the sequences of the neutral CDases from different species, all conserved serine residues were selected for site-directed mutagenesis. Of the six aligned serine residues that were mutated to alanine, only the S354A mutant lost its activity totally. Ser354 falls within a very highly conserved hexapeptide sequence GDVSPN, which itself was in the middle of a larger conserved sequence, namely NXGDVSPNXXGP/XXC. Moreover, mutations of Asp352 and Cys362 in the consensus sequence to alanine resulted in loss of activity of neutral CDase. Hence the present study identified a novel amidase sequence containing a critical serine residue that may function as a nucleophile in the hydrolytic attack on the amide bond present in ceramide.
Figures

References
-
- Michel C., van Echten-Deckert G., Rother J., Sandhoff K., Wang E., Merrill A. H., Jr Characterization of ceramide synthesis. A dihydroceramide desaturase introduces the 4,5-trans-double bond of sphingosine at the level of dihydroceramide. J. Biol. Chem. 1997;272:22432–22437. - PubMed
-
- Merrill A. H., Jr, Wang E. Enzymes of ceramide biosynthesis. Methods Enzymol. 1992;209:427–437. - PubMed
-
- Hannun Y. A., Obeid L. M. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 2002;277:25847–25850. - PubMed
-
- Sugita M., Dulaney J. T., Moser H. W. Ceramidase deficiency in Farber's disease (lipogranulomatosis) Science. 1972;178:1100–1102. - PubMed
-
- Mao C., Xu R., Bielawska A., Obeid L. M. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J. Biol. Chem. 2000;275:6876–6884. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

