Evolutionary, structural and functional relationships revealed by comparative analysis of syntenic genes in Rhizobiales
- PMID: 16229745
- PMCID: PMC1276791
- DOI: 10.1186/1471-2148-5-55
Evolutionary, structural and functional relationships revealed by comparative analysis of syntenic genes in Rhizobiales
Abstract
Background: Comparative genomics has provided valuable insights into the nature of gene sequence variation and chromosomal organization of closely related bacterial species. However, questions about the biological significance of gene order conservation, or synteny, remain open. Moreover, few comprehensive studies have been reported for rhizobial genomes.
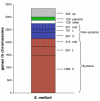
Results: We analyzed the genomic sequences of four fast growing Rhizobiales (Sinorhizobium meliloti, Agrobacterium tumefaciens, Mesorhizobium loti and Brucella melitensis). We made a comprehensive gene classification to define chromosomal orthologs, genes with homologs in other replicons such as plasmids, and those which were species-specific. About two thousand genes were predicted to be orthologs in each chromosome and about 80% of these were syntenic. A striking gene colinearity was found in pairs of organisms and a large fraction of the microsyntenic regions and operons were similar. Syntenic products showed higher identity levels than non-syntenic ones, suggesting a resistance to sequence variation due to functional constraints; also, an unusually high fraction of syntenic products contained membranal segments. Syntenic genes encode a high proportion of essential cell functions, presented a high level of functional relationships and a very low horizontal gene transfer rate. The sequence variability of the proteins can be considered the species signature in response to specific niche adaptation. Comparatively, an analysis with genomes of Enterobacteriales showed a different gene organization but gave similar results in the synteny conservation, essential role of syntenic genes and higher functional linkage among the genes of the microsyntenic regions.
Conclusion: Syntenic bacterial genes represent a commonly evolved group. They not only reveal the core chromosomal segments present in the last common ancestor and determine the metabolic characteristics shared by these microorganisms, but also show resistance to sequence variation and rearrangement, possibly due to their essential character. In Rhizobiales and Enterobacteriales, syntenic genes encode a high proportion of essential cell functions and presented a high level of functional relationships.
Figures



References
-
- Genomes Online Database http://www.genomesonline.org
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

