Symplekin and multiple other polyadenylation factors participate in 3'-end maturation of histone mRNAs
- PMID: 16230528
- PMCID: PMC1276732
- DOI: 10.1101/gad.1371105
Symplekin and multiple other polyadenylation factors participate in 3'-end maturation of histone mRNAs
Abstract
Most metazoan messenger RNAs encoding histones are cleaved, but not polyadenylated at their 3' ends. Processing in mammalian cell extracts requires the U7 small nuclear ribonucleoprotein (U7 snRNP) and an unidentified heat-labile factor (HLF). We describe the identification of a heat-sensitive protein complex whose integrity is required for histone pre-mRNA cleavage. It includes all five subunits of the cleavage and polyadenylation specificity factor (CPSF), two subunits of the cleavage stimulation factor (CstF), and symplekin. Reconstitution experiments reveal that symplekin, previously shown to be necessary for cytoplasmic poly(A) tail elongation and translational activation of mRNAs during Xenopus oocyte maturation, is the essential heat-labile component. Thus, a common molecular machinery contributes to the nuclear maturation of mRNAs both lacking and possessing poly(A), as well as to cytoplasmic poly(A) tail elongation.
Figures
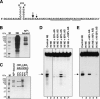


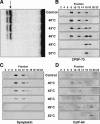
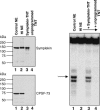

Comment in
-
Eukaryotic mRNA 3' processing: a common means to different ends.Genes Dev. 2005 Nov 1;19(21):2517-21. doi: 10.1101/gad.1378105. Genes Dev. 2005. PMID: 16264187 Review. No abstract available.
References
-
- Barabino S.M., Hübner, W., Jenny, A., Minvielle-Sebastia, L., and Keller, W. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes & Dev. 11: 1703-1716. - PubMed
-
- Barnard D.C., Ryan, K., Manley, J.L., and Richter, J.D. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641-651. - PubMed
-
- Bienroth S., Wahle, E., Suter-Crazzolara, C., and Keller, W. 1991. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J. Biol. Chem. 266: 19768-19776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
