Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism
- PMID: 16230534
- PMCID: PMC1257401
- DOI: 10.1101/gad.1352105
Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism
Abstract
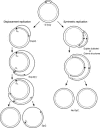
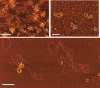
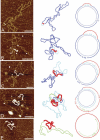
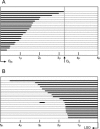
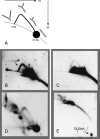
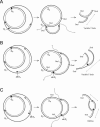
The established strand-displacement model for mammalian mitochondrial DNA (mtDNA) replication has recently been questioned in light of new data using two-dimensional (2D) agarose gel electrophoresis. It has been proposed that a synchronous, strand-coupled mode of replication occurs in tissues, thereby casting doubt on the general validity of the "orthodox," or strand-displacement model. We have examined mtDNA replicative intermediates from mouse liver using atomic force microscopy and 2D agarose gel electrophoresis in order to resolve this issue. The data provide evidence for only the orthodox, strand-displacement mode of replication and reveal the presence of additional, alternative origins of lagging light-strand mtDNA synthesis. The conditions used for 2D agarose gel analysis are favorable for branch migration of asymmetrically replicating nascent strands. These data reconcile the original displacement mode of replication with the data obtained from 2D gel analyses.
Figures

References
-
- Aloni Y. and Attardi, G. 1972. Expression of the mitochondrial genome in HeLa cells XI. Isolation and characterization of transcription complexes of mitochondrial DNA. J. Mol. Biol. 70: 363-373. - PubMed
-
- Bell L. and Byers, B. 1983. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Anal. Biochem. 130: 527-535. - PubMed
-
- Berk A.J. and Clayton, D.A. 1974. Mechanism of mitochondrial DNA replication in mouse L-cells: Asynchronous replication of strands, segregation of circular daughter molecules, aspects of topology and turnover of an initiation sequence. J. Mol. Biol. 86: 801-824. - PubMed
-
- Bogenhagen D.F. and Clayton, D.A. 2003a. The mitochondrial DNA replication bubble has not burst. Trends Biochem. Sci. 28: 357-360. - PubMed
-
- ____. 2003b. Concluding remarks: The mitochondrial DNA replication bubble has not burst. Trends Biochem. Sci. 28: 404-405. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
