Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming
- PMID: 16230535
- PMCID: PMC1257402
- DOI: 10.1101/gad.1345105
Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming
Abstract
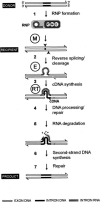
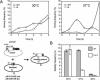
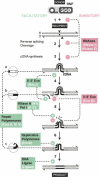
Retrohoming of group II introns occurs by a mechanism in which the intron RNA reverse splices directly into one strand of a DNA target site and is then reverse transcribed by the associated intron-encoded protein. Host repair enzymes are predicted to complete this process. Here, we screened a battery of Escherichia coli mutants defective in host functions that are potentially involved in retrohoming of the Lactococcus lactis Ll.LtrB intron. We found strong (greater than threefold) effects for several enzymes, including nucleases directed against RNA and DNA, replicative and repair polymerases, and DNA ligase. A model including the presumptive roles of these enzymes in resection of DNA, degradation of the intron RNA template, traversion of RNA-DNA junctions, and second-strand DNA synthesis is described. The completion of retrohoming is viewed as a DNA repair process, with features that may be shared by other non-LTR retroelements.
Figures


Similar articles
-
Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination.Cell. 1998 Aug 21;94(4):451-62. doi: 10.1016/s0092-8674(00)81586-x. Cell. 1998. PMID: 9727488
-
Group II intron mobility using nascent strands at DNA replication forks to prime reverse transcription.EMBO J. 2003 Sep 1;22(17):4555-65. doi: 10.1093/emboj/cdg433. EMBO J. 2003. PMID: 12941706 Free PMC article.
-
Genetic and biochemical assays reveal a key role for replication restart proteins in group II intron retrohoming.PLoS Genet. 2013 Apr;9(4):e1003469. doi: 10.1371/journal.pgen.1003469. Epub 2013 Apr 25. PLoS Genet. 2013. PMID: 23637634 Free PMC article.
-
Group II introns and expression of conjugative transfer functions in lactic acid bacteria.Antonie Van Leeuwenhoek. 1999 Jul-Nov;76(1-4):77-88. Antonie Van Leeuwenhoek. 1999. PMID: 10532373 Review.
-
Mobile group II intron targeting: applications in prokaryotes and perspectives in eukaryotes.Front Biosci. 2007 Sep 1;12:4972-85. doi: 10.2741/2442. Front Biosci. 2007. PMID: 17569624 Review.
Cited by
-
Evolution of group II introns.Mob DNA. 2015 Apr 1;6:7. doi: 10.1186/s13100-015-0037-5. eCollection 2015. Mob DNA. 2015. PMID: 25960782 Free PMC article.
-
Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions.Mob DNA. 2010 May 12;1(1):15. doi: 10.1186/1759-8753-1-15. Mob DNA. 2010. PMID: 20462415 Free PMC article.
-
Group II introns: mobile ribozymes that invade DNA.Cold Spring Harb Perspect Biol. 2011 Aug 1;3(8):a003616. doi: 10.1101/cshperspect.a003616. Cold Spring Harb Perspect Biol. 2011. PMID: 20463000 Free PMC article. Review.
-
Group II intron-based gene targeting reactions in eukaryotes.PLoS One. 2008 Sep 1;3(9):e3121. doi: 10.1371/journal.pone.0003121. PLoS One. 2008. PMID: 18769669 Free PMC article.
-
Potential role of group IIC-attC introns in integron cassette formation.J Bacteriol. 2009 Oct;191(19):6040-51. doi: 10.1128/JB.00674-09. Epub 2009 Jul 24. J Bacteriol. 2009. PMID: 19633079 Free PMC article.
References
-
- Belfort M., Derbyshire, V., Parker, M.M., Cousineau, B., and Lambowitz, A.M. 2002. Mobile introns: Pathways and proteins. In Mobile DNA II (ed. N.L. Craig et al.), pp. 761-783. ASM Press, Washington, DC.
-
- Borden A., O'Grady, P.I., Vandewiele, D., Fernández de Henestrosa, A.R., Lawrence, C.W., and Woodgate, R. 2002. Escherichia coli DNA polymerase III can replicate efficiently past a T-T cis-syn cyclobutane dimer if DNA polymerase V and the 3′ to 5′ exonuclease proofreading function encoded by dnaQ are inactivated. J. Bacteriol. 184: 2674-2681. - PMC - PubMed
-
- Brescia C.C., Kaw, M.K., and Sledjeski, D.D. 2004. The DNA binding protein H-NS binds to and alters the stability of RNA in vitro and in vivo. J. Mol. Biol. 339: 505-514. - PubMed
-
- Coros C.J., Landthaler, M., Piazza, C.L., Beauregard, A., Esposito, D., Perutka, J., Lambowitz, A.M., and Belfort, M. 2005. Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol. Microbiol. 56: 509-524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
