Differential recruitment of pre-mRNA splicing factors to alternatively spliced transcripts in vivo
- PMID: 16231974
- PMCID: PMC1262628
- DOI: 10.1371/journal.pbio.0030374
Differential recruitment of pre-mRNA splicing factors to alternatively spliced transcripts in vivo
Abstract
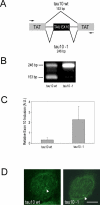
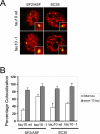
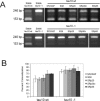
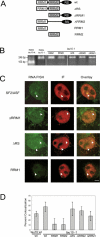
Alternative splicing in mammalian cells has been suggested to be largely controlled by combinatorial binding of basal splicing factors to pre-mRNA templates. This model predicts that distinct sets of pre-mRNA splicing factors are associated with alternatively spliced transcripts. However, no experimental evidence for differential recruitment of splicing factors to transcripts with distinct splicing fates is available. Here we have used quantitative single-cell imaging to test this key prediction in vivo. We show that distinct combinations of splicing factors are recruited to sites of alternatively spliced transcripts in intact cells. While a subset of serine/arginine protein splicing factors, including SF2/ASF, SC35, and SRp20, is efficiently recruited to the tau gene when exon 10 is included, these factors are less frequently associated with tau transcription sites when exon 10 is excluded. In contrast, the frequency of recruitment of several other splicing factors is independent of splicing outcome. Mutation analysis of SF2/ASF shows that both protein-protein as well as protein-RNA interactions are required for differential recruitment. The differential behavior of the various splicing factors provides the basis for combinatorial occupancy at pre-mRNAs. These observations represent the first in vivo evidence for differential association of pre-mRNA splicing factors with alternatively spliced transcripts. They confirm a key prediction of a stochastic model of alternative splicing, in which distinct combinatorial sets of generic pre-mRNA splicing factors contribute to splicing outcome.
Figures


References
-
- Sanford JR, Caceres JF. Pre-mRNA splicing: Life at the centre of the central dogma. J Cell Sci. 2004;117:6261–6263. - PubMed
-
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. - PubMed
-
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

