The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales
- PMID: 16236178
- PMCID: PMC1277820
- DOI: 10.1186/1741-7007-3-22
The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales
Abstract
Background: The Streptophyta comprise all land plants and six monophyletic groups of charophycean green algae. Phylogenetic analyses of four genes from three cellular compartments support the following branching order for these algal lineages: Mesostigmatales, Chlorokybales, Klebsormidiales, Zygnematales, Coleochaetales and Charales, with the last lineage being sister to land plants. Comparative analyses of the Mesostigma viride (Mesostigmatales) and land plant chloroplast genome sequences revealed that this genome experienced many gene losses, intron insertions and gene rearrangements during the evolution of charophyceans. On the other hand, the chloroplast genome of Chaetosphaeridium globosum (Coleochaetales) is highly similar to its land plant counterparts in terms of gene content, intron composition and gene order, indicating that most of the features characteristic of land plant chloroplast DNA (cpDNA) were acquired from charophycean green algae. To gain further insight into when the highly conservative pattern displayed by land plant cpDNAs originated in the Streptophyta, we have determined the cpDNA sequences of the distantly related zygnematalean algae Staurastrum punctulatum and Zygnema circumcarinatum.
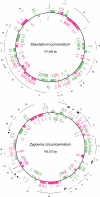
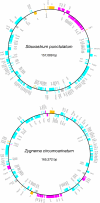
Results: The 157,089 bp Staurastrum and 165,372 bp Zygnema cpDNAs encode 121 and 125 genes, respectively. Although both cpDNAs lack an rRNA-encoding inverted repeat (IR), they are substantially larger than Chaetosphaeridium and land plant cpDNAs. This increased size is explained by the expansion of intergenic spacers and introns. The Staurastrum and Zygnema genomes differ extensively from one another and from their streptophyte counterparts at the level of gene order, with the Staurastrum genome more closely resembling its land plant counterparts than does Zygnema cpDNA. Many intergenic regions in Zygnema cpDNA harbor tandem repeats. The introns in both Staurastrum (8 introns) and Zygnema (13 introns) cpDNAs represent subsets of those found in land plant cpDNAs. They represent 16 distinct insertion sites, only five of which are shared by the two zygnematalean genomes. Three of these insertions sites have not been identified in Chaetosphaeridium cpDNA.
Conclusion: The chloroplast genome experienced substantial changes in overall structure, gene order, and intron content during the evolution of the Zygnematales. Most of the features considered earlier as typical of land plant cpDNAs probably originated before the emergence of the Zygnematales and Coleochaetales.
Figures



Similar articles
-
An unexpectedly large and loosely packed mitochondrial genome in the charophycean green alga Chlorokybus atmophyticus.BMC Genomics. 2007 May 30;8:137. doi: 10.1186/1471-2164-8-137. BMC Genomics. 2007. PMID: 17537252 Free PMC article.
-
The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants.Mol Biol Evol. 2006 Jun;23(6):1324-38. doi: 10.1093/molbev/msk018. Epub 2006 Apr 12. Mol Biol Evol. 2006. PMID: 16611644
-
The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants.Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11275-80. doi: 10.1073/pnas.162203299. Epub 2002 Aug 2. Proc Natl Acad Sci U S A. 2002. PMID: 12161560 Free PMC article.
-
Green algae and the evolution of land plants: inferences from nuclear-encoded rRNA gene sequences.Biosystems. 1992;28(1-3):127-37. doi: 10.1016/0303-2647(92)90015-q. Biosystems. 1992. PMID: 1292658 Review.
-
Desiccation tolerance in streptophyte algae and the algae to land plant transition: evolution of LEA and MIP protein families within the Viridiplantae.J Exp Bot. 2020 Jun 11;71(11):3270-3278. doi: 10.1093/jxb/eraa105. J Exp Bot. 2020. PMID: 32107542 Free PMC article. Review.
Cited by
-
Complex RNA metabolism in the chloroplast: an update on the psbB operon.Planta. 2013 Feb;237(2):441-9. doi: 10.1007/s00425-012-1782-z. Epub 2012 Oct 13. Planta. 2013. PMID: 23065055 Free PMC article. Review.
-
An unexpectedly large and loosely packed mitochondrial genome in the charophycean green alga Chlorokybus atmophyticus.BMC Genomics. 2007 May 30;8:137. doi: 10.1186/1471-2164-8-137. BMC Genomics. 2007. PMID: 17537252 Free PMC article.
-
The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured.BMC Biol. 2006 Apr 21;4:12. doi: 10.1186/1741-7007-4-12. BMC Biol. 2006. PMID: 16630350 Free PMC article.
-
The chloroplast sulfate transport system in the green alga Chlamydomonas reinhardtii.Planta. 2008 Nov;228(6):951-61. doi: 10.1007/s00425-008-0795-0. Epub 2008 Aug 6. Planta. 2008. PMID: 18682979
-
Genomes of multicellular algal sisters to land plants illuminate signaling network evolution.Nat Genet. 2024 May;56(5):1018-1031. doi: 10.1038/s41588-024-01737-3. Epub 2024 May 1. Nat Genet. 2024. PMID: 38693345 Free PMC article.
References
-
- Kenrick P, Crane PR. The origin and early evolution of plants on land. Nature. 1997;389:33–39. doi: 10.1038/37918. - DOI
-
- Sanderson MJ, Thorne JL, Wikstrom N, Bremer K. Molecular evidence on plant divergence times. Am J Bot. 2004;91:1656–1665. - PubMed
-
- Lewis LA, McCourt RM. Green algae and the origin of land plants. Am J Bot. 2004;91:1535–1556. - PubMed
-
- Bremer K, Humphries CJ, Mishler BD, Churchill SP. On cladistic relationships in green plants. Taxon. 1987;36:339–349.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

