Low-frequency normal mode in DNA HhaI methyltransferase and motions of residues involved in the base flipping
- PMID: 16236720
- PMCID: PMC1283451
- DOI: 10.1073/pnas.0507913102
Low-frequency normal mode in DNA HhaI methyltransferase and motions of residues involved in the base flipping
Abstract
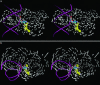
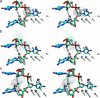
The results of normal-mode analyses are in accord with the proposal that a low-frequency motion of the HhaI methyltransferase enzyme is responsible for base flipping in bound DNA. The vectors of the low-frequency normal mode of residues Ser-85 and Ile-86 point directly to the phosphate and ribose moieties of the DNA backbone near the target base in position to rotate the dihedral angles and flip the base out of the DNA duplex. The vector of residue Gln-237 on the major groove is in the proper orientation to assist base separation. Our results favor the major groove pathway and the protein active process in base flipping.
Figures
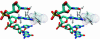

Similar articles
-
Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase.Nucleic Acids Res. 2004 Jul 23;32(13):3877-86. doi: 10.1093/nar/gkh701. Print 2004. Nucleic Acids Res. 2004. PMID: 15273274 Free PMC article.
-
Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase.Proc Natl Acad Sci U S A. 2003 Jan 7;100(1):68-73. doi: 10.1073/pnas.0135427100. Epub 2002 Dec 27. Proc Natl Acad Sci U S A. 2003. PMID: 12506195 Free PMC article.
-
Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.Nat Struct Biol. 1998 Oct;5(10):872-7. doi: 10.1038/2312. Nat Struct Biol. 1998. PMID: 9783745
-
Finding a basis for flipping bases.Structure. 1996 Jun 15;4(6):639-45. doi: 10.1016/s0969-2126(96)00068-8. Structure. 1996. PMID: 8805547 Review.
-
Atomistic view of base flipping in DNA.Philos Trans A Math Phys Eng Sci. 2004 Jul 15;362(1820):1439-60. doi: 10.1098/rsta.2004.1383. Philos Trans A Math Phys Eng Sci. 2004. PMID: 15306460 Review.
Cited by
-
Base Dynamics in the HhaI Protein Binding Site.J Phys Chem B. 2023 Aug 24;127(33):7266-7275. doi: 10.1021/acs.jpcb.3c03687. Epub 2023 Aug 10. J Phys Chem B. 2023. PMID: 37561575 Free PMC article.
-
Hidden Conformation Events in DNA Base Extrusions: A Generalized Ensemble Path Optimization and Equilibrium Simulation Study.J Chem Theory Comput. 2013 Aug 13;9(8):10.1021/ct400198q. doi: 10.1021/ct400198q. J Chem Theory Comput. 2013. PMID: 24250279 Free PMC article.
-
A molecular dynamics study of slow base flipping in DNA using conformational flooding.Biophys J. 2007 Aug 1;93(3):770-86. doi: 10.1529/biophysj.106.091751. Epub 2007 May 11. Biophys J. 2007. PMID: 17496048 Free PMC article.
-
Coupling between catalytic loop motions and enzyme global dynamics.PLoS Comput Biol. 2012;8(9):e1002705. doi: 10.1371/journal.pcbi.1002705. Epub 2012 Sep 27. PLoS Comput Biol. 2012. PMID: 23028297 Free PMC article.
References
-
- Doerfler, W. (1983) Annu. Rev. Biochem. 52, 93–124. - PubMed
-
- Wu, J. C. & Santi, D. V. (1987) J. Biol. Chem. 262, 4778–4786. - PubMed
-
- Roberts, R. J., Myers, P. A., Morrison, A. & Murray, K. (1976) J. Mol. Biol. 103, 199–208. - PubMed
-
- Mann, M. B. & Smith, H. O. (1979) in Proceedings of the Conference on Transmethylation, eds. Usdin, E., Borchardt, R. T. & Creveling, C. R. (Elsevier, New York), pp. 483–492.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

