Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome
- PMID: 16236728
- PMCID: PMC1276048
- DOI: 10.1073/pnas.0503868102
Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome
Abstract
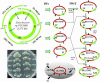
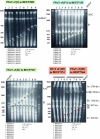
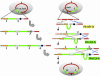
Cloning the whole 3.5-megabase (Mb) genome of the photosynthetic bacterium Synechocystis PCC6803 into the 4.2-Mb genome of the mesophilic bacterium Bacillus subtilis 168 resulted in a 7.7-Mb composite genome. We succeeded in such unprecedented large-size cloning by progressively assembling and editing contiguous DNA regions that cover the entire Synechocystis genome. The strain containing the two sets of genome grew only in the B. subtilis culture medium where all of the cloning procedures were carried out. The high structural stability of the cloned Synechocystis genome was closely associated with the symmetry of the bacterial genome structure of the DNA replication origin (oriC) and its termination (terC) and the exclusivity of Synechocystis ribosomal RNA operon genes (rrnA and rrnB). Given the significant diversity in genome structure observed upon horizontal DNA transfer in nature, our stable laboratory-generated composite genome raised fundamental questions concerning two complete genomes in one cell. Our megasize DNA cloning method, designated megacloning, may be generally applicable to other genomes or genome loci of free-living organisms.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

