Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II
- PMID: 16237000
- PMCID: PMC1272990
- DOI: 10.1128/JB.187.21.7167-7175.2005
Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II
Abstract
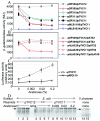
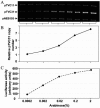
Replication initiator proteins in bacteria not only allow DNA replication but also often regulate the rate of replication initiation as well. The regulation is mediated by limiting the synthesis or availability of initiator proteins. The applicability of this principle is demonstrated here for RctB, the replication initiator for the smaller of the two chromosomes of Vibrio cholerae. A strong promoter for the rctB gene named rctBp was identified and found to be autoregulated in Escherichia coli. Promoter activity was lower in V. cholerae than in E. coli, and a part of this reduction is likely to be due to autorepression. Sequences upstream of rctBp, implicated earlier in replication control, enhanced the repression. The action of the upstream sequences required that they be present in cis, implying long-range interactions in the control of the promoter activity. A second gene specific for chromosome II replication, rctA, reduced rctB translation, most likely by antisense RNA control. Finally, optimal rctBp activity was found to be dependent on Dam. Increasing RctB in trans increased the copy number of a miniplasmid carrying oriCII(VC), implying that RctB can be rate limiting for chromosome II replication. The multiple modes of control on RctB are expected to reduce fluctuations in the initiator concentration and thereby help maintain chromosome copy number homeostasis.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

