Functional consequences of genome evolution in Listeria monocytogenes: the lmo0423 and lmo0422 genes encode sigmaC and LstR, a lineage II-specific heat shock system
- PMID: 16237008
- PMCID: PMC1272999
- DOI: 10.1128/JB.187.21.7243-7253.2005
Functional consequences of genome evolution in Listeria monocytogenes: the lmo0423 and lmo0422 genes encode sigmaC and LstR, a lineage II-specific heat shock system
Abstract
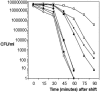
Listeria monocytogenes strains belonging to phylogenetic lineage II (serotypes 1/2a, 1/2c, and 3a) carry a lineage-specific genome segment encoding a putative sigma subunit of RNA polymerase (lmo0423, herein referred to as sigC), a gene of unknown function (lmo0422) similar to the padR family of regulators, and a gene that is similar to the rodA-ftsW family of cell wall morphology genes (lmo0421). To understand the function of this set of genes, their expression patterns and the effects of null mutations in the lineage II L. monocytogenes strain 10403S were examined. The data are consistent with the three genes comprising an operon (the sigC operon) that is highly induced by temperature upshift. The operon is transcribed from three different promoters, the proximal of which (P1) depends upon sigC itself. Null mutations in sigC or lmo0422 increase the death rate at lethal temperatures and cause loss of thermal adaptive response, whereas the lmo0421 mutation causes only a loss of the adaptive response component. Only the sigC mutation affects transcription from the P1 promoter, whereas ectopic expression of lmo0422 from the P(SPAC) promoter complements the individual lmo0422 and sigC null mutations, showing that lmo0422 is the actual thermal resistance regulator or effector while sigC provides a mechanism for temperature-dependent transcription of lmo0422 from P1. Our genetic and phylogenetic analyses are consistent with lmo0422-renamed lstR (for lineage-specific thermal regulator)-and sigC comprising a system of thermal resistance that was ancestral to the genus Listeria and was subsequently lost during divergence of the lineage I L. monocytogenes population.
Figures



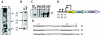

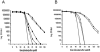


Similar articles
-
Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set.Mol Microbiol. 2004 Apr;52(1):25-38. doi: 10.1111/j.1365-2958.2003.03958.x. Mol Microbiol. 2004. PMID: 15049808
-
Contributions of six lineage-specific internalin-like genes to invasion efficiency of Listeria monocytogenes.Foodborne Pathog Dis. 2009 Jan-Feb;6(1):57-70. doi: 10.1089/fpd.2008.0140. Foodborne Pathog Dis. 2009. PMID: 19014275
-
Cloning, sequencing, and transcriptional analysis of the dnaK heat shock operon of Listeria monocytogenes.Cell Stress Chaperones. 2000 Jan;5(1):21-9. doi: 10.1043/1355-8145(2000)005<0021:CSATAO>2.0.CO;2. Cell Stress Chaperones. 2000. PMID: 10701836 Free PMC article.
-
Identification and analysis of the osmotolerance associated genes in Listeria monocytogenes.Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008 Sep;25(9):1089-94. doi: 10.1080/02652030802056634. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008. PMID: 19238619 Review.
-
Regulation of nitrogen fixation genes.Annu Rev Genet. 1986;20:567-91. doi: 10.1146/annurev.ge.20.120186.003031. Annu Rev Genet. 1986. PMID: 3545064 Review. No abstract available.
Cited by
-
Regulatory network features in Listeria monocytogenes-changing the way we talk.Front Cell Infect Microbiol. 2014 Feb 14;4:14. doi: 10.3389/fcimb.2014.00014. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 24592357 Free PMC article. Review.
-
Genetic and biochemical analysis of PadR-padC promoter interactions during the phenolic acid stress response in Bacillus subtilis 168.J Bacteriol. 2011 Aug;193(16):4180-91. doi: 10.1128/JB.00385-11. Epub 2011 Jun 17. J Bacteriol. 2011. PMID: 21685295 Free PMC article.
-
Modulation of stress and virulence in Listeria monocytogenes.Trends Microbiol. 2008 Aug;16(8):388-96. doi: 10.1016/j.tim.2008.05.006. Epub 2008 Jul 9. Trends Microbiol. 2008. PMID: 18619843 Free PMC article. Review.
-
PadR-type repressors controlling production of a non-canonical FtsW/RodA homologue and other trans-membrane proteins.Sci Rep. 2019 Jul 11;9(1):10023. doi: 10.1038/s41598-019-46347-w. Sci Rep. 2019. PMID: 31296881 Free PMC article.
-
The Analysis of Field Strains Isolated From Food, Animal and Clinical Sources Uncovers Natural Mutations in Listeria monocytogenes Nisin Resistance Genes.Front Microbiol. 2020 Oct 6;11:549531. doi: 10.3389/fmicb.2020.549531. eCollection 2020. Front Microbiol. 2020. PMID: 33123101 Free PMC article.
References
-
- Arous, S., C. Buchrieser, P. Folio, P. Glaser, A. Namane, M. Hebraud, and Y. Hechard. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581-1590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

