Chlamydial GroEL autoregulates its own expression through direct interactions with the HrcA repressor protein
- PMID: 16237037
- PMCID: PMC1272993
- DOI: 10.1128/JB.187.21.7535-7542.2005
Chlamydial GroEL autoregulates its own expression through direct interactions with the HrcA repressor protein
Abstract
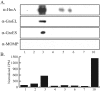
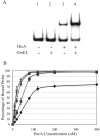
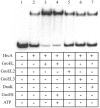
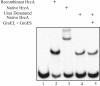
In the pathogenic bacterium Chlamydia trachomatis, a transcriptional repressor, HrcA, regulates the major heat shock operons, dnaK and groE. Cellular stress causes a transient increase in transcription of these heat shock operons through relief of HrcA-mediated repression, but the pathway leading to derepression is unclear. Elevated temperature alone is not sufficient, and it is hypothesized that additional chlamydial factors play a role. We used DNA affinity chromatography to purify proteins that interact with HrcA in vivo and identified a higher-order complex consisting of HrcA, GroEL, and GroES. This endogenous HrcA complex migrated differently than recombinant HrcA, but the complex could be disrupted, releasing native HrcA that resembled recombinant HrcA. In in vitro assays, GroEL increased the ability of HrcA to bind to the CIRCE operator and to repress transcription. Other chlamydial heat shock proteins, including the two additional GroEL paralogs present in all chlamydial species, did not modulate HrcA activity.
Figures


Similar articles
-
Stress response gene regulation in Chlamydia is dependent on HrcA-CIRCE interactions.J Bacteriol. 2004 Jun;186(11):3384-91. doi: 10.1128/JB.186.11.3384-3391.2004. J Bacteriol. 2004. PMID: 15150223 Free PMC article.
-
A Chlamydia-specific C-terminal region of the stress response regulator HrcA modulates its repressor activity.J Bacteriol. 2011 Dec;193(23):6733-41. doi: 10.1128/JB.05792-11. Epub 2011 Sep 30. J Bacteriol. 2011. PMID: 21965565 Free PMC article.
-
The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis.EMBO J. 1997 Aug 1;16(15):4579-90. doi: 10.1093/emboj/16.15.4579. EMBO J. 1997. PMID: 9303302 Free PMC article.
-
Negative regulation of the heat shock response in Streptomyces.Arch Microbiol. 2001 Oct;176(4):237-42. doi: 10.1007/s002030100321. Arch Microbiol. 2001. PMID: 11685367 Review.
-
The Escherichia coli groE chaperonins.Semin Cell Biol. 1990 Feb;1(1):19-25. Semin Cell Biol. 1990. PMID: 1983267 Review.
Cited by
-
Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival.BMC Microbiol. 2006 Dec 18;6:102. doi: 10.1186/1471-2180-6-102. BMC Microbiol. 2006. PMID: 17176467 Free PMC article.
-
Identification and functional analysis of CT069 as a novel transcriptional regulator in Chlamydia.J Bacteriol. 2011 Nov;193(22):6123-31. doi: 10.1128/JB.05976-11. Epub 2011 Sep 9. J Bacteriol. 2011. PMID: 21908669 Free PMC article.
-
Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis.J Bacteriol. 2006 Jun;188(12):4236-43. doi: 10.1128/JB.01660-05. J Bacteriol. 2006. PMID: 16740930 Free PMC article.
-
Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements.J Bacteriol. 2007 Jan;189(1):198-206. doi: 10.1128/JB.01034-06. Epub 2006 Oct 20. J Bacteriol. 2007. PMID: 17056752 Free PMC article.
-
Differential effects of DNA supercoiling on Chlamydia early promoters correlate with expression patterns in midcycle.J Bacteriol. 2012 Jun;194(12):3109-15. doi: 10.1128/JB.00242-12. Epub 2012 Apr 13. J Bacteriol. 2012. PMID: 22505684 Free PMC article.
References
-
- Ausubel, F. M., R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, New York, N.Y.
-
- Beatty, W., G. I. Byrne, and R. P. Morrison. 1994. Repeated and persistent infection with Chlamydia and the development of chronic inflammation and disease. Trends Microbiol. 2:94-98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

