Atomic force microscopy shows that vaccinia topoisomerase IB generates filaments on DNA in a cooperative fashion
- PMID: 16237128
- PMCID: PMC1258176
- DOI: 10.1093/nar/gki906
Atomic force microscopy shows that vaccinia topoisomerase IB generates filaments on DNA in a cooperative fashion
Abstract
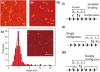
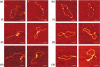
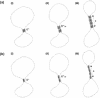
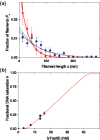
Type IB DNA topoisomerases cleave and rejoin one strand of the DNA duplex, allowing for the removal of supercoils generated during replication and transcription. In addition, electron microscopy of cellular and viral TopIB-DNA complexes has suggested that the enzyme promotes long-range DNA-DNA crossovers and synapses. Here, we have used the atomic force microscope to visualize and quantify the interaction between vaccinia topoisomerase IB (vTopIB) and DNA. vTopIB was found to form filaments on nicked-circular DNA by intramolecular synapsis of two segments of a single DNA molecule. Measuring the filament length as a function of protein concentration showed that synapsis is a highly cooperative process. At high protein:DNA ratios, synapses between distinct DNA molecules were observed, which led to the formation of large vTopIB-induced DNA clusters. These clusters were observed in the presence of Mg2+, Ca2+ or Mn2+, suggesting that the formation of intermolecular vTopIB-mediated DNA synapsis is favored by screening of the DNA charge.
Figures


Similar articles
-
Intramolecular synapsis of duplex DNA by vaccinia topoisomerase.EMBO J. 1997 Nov 3;16(21):6584-9. doi: 10.1093/emboj/16.21.6584. EMBO J. 1997. PMID: 9351838 Free PMC article.
-
Characterization of DNA Binding by the Isolated N-Terminal Domain of Vaccinia Virus DNA Topoisomerase IB.Biochemistry. 2017 Jul 5;56(26):3307-3317. doi: 10.1021/acs.biochem.7b00042. Epub 2017 Jun 19. Biochemistry. 2017. PMID: 28570045 Free PMC article.
-
Expression and Purification of Vaccinia Virus DNA Topoisomerase IB Produced in the Silkworm-Baculovirus Expression System.Mol Biotechnol. 2019 Aug;61(8):622-630. doi: 10.1007/s12033-019-00184-4. Mol Biotechnol. 2019. PMID: 31165966
-
Teaching a new dog old tricks?Structure. 1998 May 15;6(5):543-8. doi: 10.1016/s0969-2126(98)00056-2. Structure. 1998. PMID: 9634692 Review.
-
Conserved themes but novel activities in recombinases and topoisomerases.Cell. 1998 Apr 17;93(2):149-52. doi: 10.1016/s0092-8674(00)81566-4. Cell. 1998. PMID: 9568707 Review. No abstract available.
Cited by
-
A highly processive topoisomerase I: studies at the single-molecule level.Nucleic Acids Res. 2014 Jul;42(12):7935-46. doi: 10.1093/nar/gku494. Epub 2014 May 31. Nucleic Acids Res. 2014. PMID: 24880688 Free PMC article.
-
Variola type IB DNA topoisomerase: DNA binding and supercoil unwinding using engineered DNA minicircles.Biochemistry. 2014 Jul 8;53(26):4302-15. doi: 10.1021/bi500571q. Epub 2014 Jun 26. Biochemistry. 2014. PMID: 24945825 Free PMC article.
-
Simulations of DNA topoisomerase 1B bound to supercoiled DNA reveal changes in the flexibility pattern of the enzyme and a secondary protein-DNA binding site.Nucleic Acids Res. 2014 Aug;42(14):9304-12. doi: 10.1093/nar/gku654. Epub 2014 Jul 23. Nucleic Acids Res. 2014. PMID: 25056319 Free PMC article.
-
Structures of topoisomerase V in complex with DNA reveal unusual DNA-binding mode and novel relaxation mechanism.Elife. 2022 Aug 15;11:e72702. doi: 10.7554/eLife.72702. Elife. 2022. PMID: 35969036 Free PMC article.
-
Topoisomerase IB-DNA interactions: X marks the spot.Structure. 2010 Jun 9;18(6):661-3. doi: 10.1016/j.str.2010.05.002. Structure. 2010. PMID: 20541502 Free PMC article.
References
-
- Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. - PubMed
-
- Cheng C., Kussie P., Pavletich N., Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. - PubMed
-
- Shuman S., Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J. Biol. Chem. 1990;265:17826–17836. - PubMed
-
- Shuman S. Site-specific interaction of vaccinia virus topoisomerase I with duplex DNA. Minimal DNA substrate for strand cleavage in vitro. J. Biol. Chem. 1991;266:11372–11379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

