Assay validation for identification of hereditary nonpolyposis colon cancer-causing mutations in mismatch repair genes MLH1, MSH2, and MSH6
- PMID: 16237223
- PMCID: PMC1888496
- DOI: 10.1016/S1525-1578(10)60584-3
Assay validation for identification of hereditary nonpolyposis colon cancer-causing mutations in mismatch repair genes MLH1, MSH2, and MSH6
Abstract
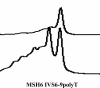
Hereditary nonpolyposis colon cancer (HNPCC, Online Mendelian Inheritance in Man (OMIM) 114500) is an autosomal dominant disorder that is genetically heterogeneous because of underlying mutations in mismatch repair genes, primarily MLH1, MSH2, and MSH6. One challenge to correctly diagnosing HNPCC is that the large size of the causative genes makes identification of mutations both labor intensive and expensive. We evaluated the usefulness of denaturing high performance liquid chromatography (DHPLC) for scanning mismatch repair genes (MLH1, MSH2, and MSH6) for point mutations, small deletions, and insertions. Our assay consisted of 51 sets of primers designed to amplify all exons of these genes. All polymerase chain reaction reactions were amplified simultaneously using the same reaction conditions in a 96-well format. The amplified products were analyzed by DHPLC across a range of optimum temperatures for partial fragment denaturation based on the melting profile of each specific fragment. DNA specimens from 23 previously studied HNPCC patients were analyzed by DHPLC, and all mutations were correctly identified and confirmed by sequence analysis. Here, we present our validation studies of the DHPLC platform for HNPCC mutation analysis and compare its merits with other scanning technologies. This approach provides greater sensitivity and more directed molecular analysis for clinical testing in HNPCC.
Figures


Similar articles
-
Genetic testing for hereditary nonpolyposis colorectal cancer (HNPCC).Curr Protoc Hum Genet. 2009 Apr;Chapter 10:Unit 10.12. doi: 10.1002/0471142905.hg1012s61. Curr Protoc Hum Genet. 2009. PMID: 19360696
-
Two mismatch repair gene mutations found in a colon cancer patient--which one is pathogenic?Hum Genet. 2003 Feb;112(2):105-9. doi: 10.1007/s00439-002-0866-4. Epub 2002 Nov 21. Hum Genet. 2003. PMID: 12522549
-
Multiplex SNaPshot genotyping for detecting loss of heterozygosity in the mismatch-repair genes MLH1 and MSH2 in microsatellite-unstable tumors.Clin Chem. 2008 Nov;54(11):1844-54. doi: 10.1373/clinchem.2008.108902. Epub 2008 Sep 4. Clin Chem. 2008. PMID: 18772310
-
Hereditary nonpolyposis colorectal cancer: diagnostic strategies and their implications.Evid Rep Technol Assess (Full Rep). 2007 May;(150):1-180. Evid Rep Technol Assess (Full Rep). 2007. PMID: 17764220 Free PMC article. Review.
-
Do MSH6 mutations contribute to double primary cancers of the colorectum and endometrium?Hum Genet. 2000 Dec;107(6):623-9. doi: 10.1007/s004390000417. Hum Genet. 2000. PMID: 11153917 Review.
Cited by
-
Suspected Lynch syndrome associated MSH6 variants: A functional assay to determine their pathogenicity.PLoS Genet. 2017 May 22;13(5):e1006765. doi: 10.1371/journal.pgen.1006765. eCollection 2017 May. PLoS Genet. 2017. PMID: 28531214 Free PMC article.
-
Targeted sequencing of genes associated with the mismatch repair pathway in patients with endometrial cancer.PLoS One. 2020 Jul 7;15(7):e0235613. doi: 10.1371/journal.pone.0235613. eCollection 2020. PLoS One. 2020. PMID: 32634176 Free PMC article.
-
Microsatellite instability in pediatric high grade glioma is associated with genomic profile and differential target gene inactivation.PLoS One. 2011;6(5):e20588. doi: 10.1371/journal.pone.0020588. Epub 2011 May 26. PLoS One. 2011. PMID: 21637783 Free PMC article.
-
Characterization of two Ashkenazi Jewish founder mutations in MSH6 gene causing Lynch syndrome.Clin Genet. 2011 Jun;79(6):512-22. doi: 10.1111/j.1399-0004.2010.01594.x. Epub 2010 Dec 14. Clin Genet. 2011. PMID: 21155762 Free PMC article.
-
Mutational analysis of thyroid transcription factor-1 gene (TTF-1) in lung carcinomas.In Vitro Cell Dev Biol Anim. 2008 Jan-Feb;44(1-2):17-25. doi: 10.1007/s11626-007-9062-0. Epub 2007 Dec 11. In Vitro Cell Dev Biol Anim. 2008. PMID: 18071837
References
-
- Burt RW. Familial risk and colorectal cancer. Gastroenterol Clin North Am. 1996;25:793–803. - PubMed
-
- Burt RW. Familial association. Adv Exp Med Biol. 1999;470:99–104. - PubMed
-
- Coughlin SS, Miller DS. Public health perspectives on testing for colorectal cancer susceptibility genes. Am J Prev Med. 1999;16:99–104. - PubMed
-
- Cederquist K, Golovleva I, Emanuelsson M, Stenling R, Gronberg H. A population based cohort study of patients with multiple colon and endometrial cancer: correlation of microsatellite instability (MSI) status, age at diagnosis and cancer risk. Int J Cancer. 2001;91:486–491. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

