Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin
- PMID: 16239142
- PMCID: PMC2039754
- DOI: 10.1016/j.cell.2005.10.002
Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin
Erratum in
- Cell. 2008 Jul 11;134(1):188
Abstract
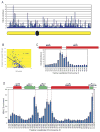
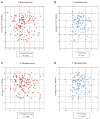
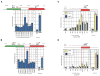
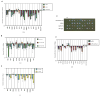
In S. cerevisiae, histone variant H2A.Z is deposited in euchromatin at the flanks of silent heterochromatin to prevent its ectopic spread. We show that H2A.Z nucleosomes are found at promoter regions of nearly all genes in euchromatin. They generally occur as two positioned nucleosomes that flank a nucleosome-free region (NFR) that contains the transcription start site. Astonishingly, enrichment at 5' ends is observed not only at actively transcribed genes but also at inactive loci. Mutagenesis of a typical promoter revealed a 22 bp segment of DNA sufficient to program formation of a NFR flanked by two H2A.Z nucleosomes. This segment contains a binding site of the Myb-related protein Reb1 and an adjacent dT:dA tract. Efficient deposition of H2A.Z is further promoted by a specific pattern of histone H3 and H4 tail acetylation and the bromodomain protein Bdf1, a component of the Swr1 remodeling complex that deposits H2A.Z.
Figures


References
-
- Angermayr M, Oechsner U, Bandlow W. Reb1p-dependent DNA bending effects nucleosome positioning and constitutive transcription at the yeast profilin promoter. J Biol Chem. 2003;278:17918–17926. - PubMed
-
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

