Cell cycle features of primate embryonic stem cells
- PMID: 16239321
- PMCID: PMC1934406
- DOI: 10.1634/stemcells.2005-0194
Cell cycle features of primate embryonic stem cells
Abstract
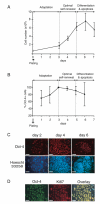
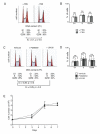
Using flow cytometry measurements combined with quantitative analysis of cell cycle kinetics, we show that rhesus monkey embryonic stem cells (ESCs) are characterized by an extremely rapid transit through the G1 phase, which accounts for 15% of the total cell cycle duration. Monkey ESCs exhibit a non-phasic expression of cyclin E, which is detected during all phases of the cell cycle, and do not growth-arrest in G1 after gamma-irradiation, reflecting the absence of a G1 checkpoint. Serum deprivation or pharmacological inhibition of mitogen-activated protein kinase kinase (MEK) did not result in any alteration in the cell cycle distribution, indicating that ESC growth does not rely on mitogenic signals transduced by the Ras/Raf/MEK pathway. Taken together, these data indicate that rhesus monkey ESCs, like their murine counterparts, exhibit unusual cell cycle features in which cell cycle control mechanisms operating during the G1 phase are reduced or absent.
Figures





References
-
- Savatier P, Lapillonne H, van Grunsven LA, et al. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12:309–309. - PubMed
-
- Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

