Merozoite surface antigen 2 proteins of Babesia bovis vaccine breakthrough isolates contain a unique hypervariable region composed of degenerate repeats
- PMID: 16239512
- PMCID: PMC1273907
- DOI: 10.1128/IAI.73.11.7180-7189.2005
Merozoite surface antigen 2 proteins of Babesia bovis vaccine breakthrough isolates contain a unique hypervariable region composed of degenerate repeats
Abstract
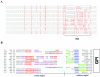
The merozoite surface antigen 2 (MSA-2) proteins of Babesia bovis are members of the variable merozoite surface antigen (VMSA) family that have been implicated in erythrocyte invasion and are important targets for antibody-mediated blocking of invasion. Extensive sequence variation in another VMSA member, MSA-1, has been shown in all vaccine breakthrough isolates. To test the hypothesis that the msa-2 genes of vaccine breakthrough isolates would also encode a diverse set of proteins, the complete msa-2 locus was characterized from 12 Australian B. bovis strains and isolates, including two vaccine strains and eight vaccine breakthrough isolates, and compared to the loci in previously and newly characterized American strains. In contrast to American strains, the msa-2 loci of all Australian strains and isolates examined contain, in addition to msa-2c, only a solitary gene (designated msa-2a/b) closely related to American strain msa-2a and msa-2b. Nevertheless, the proteins encoded by these genes are quite diverse both between and within geographic regions and harbor evidence of genetic exchange among other VMSA family members, including msa-1. Moreover, all but one of the Australian breakthrough isolate MSA-2a/b proteins is markedly different from the vaccine strain from which immune escape occurred, consistent with their role in strain-specific protective immunity. The densest distribution of polymorphisms occurs in a hypervariable region (HVR) within the carboxy third of the molecule that is highly proline rich. Variation in length and content of the HVR is primarily attributable to differences in the order and number of degenerate nucleotide repeats encoding three motifs of unknown function.
Figures




References
-
- Anziani, O. S., A. A. Guglielmone, A. A. Abdala, D. H. Aguirre, and A. J. Mangold. 1993. Acquired protection in Frisian steers with Babesia bovis attenuated vaccine. Rev. Med. Vet. 74:47-49.
-
- Ayala, F. J., A. A. Escalante, and S. M. Rich. 1999. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum. Parasitologia 41:55-68. - PubMed
-
- Bock, R. E., A. J. de Vos, T. G. Kingston, I. A. Shiels, and R. J. Dalgliesh. 1992. Investigations of breakdowns in protection provided by living Babesia bovis vaccine. Vet. Parasitol. 43:45-56. - PubMed
-
- Bock, R. E., A. J. de Vos, A. Lew, T. G. Kingston, and I. R. Fraser. 1995. Studies on failure of T strain live Babesia bovis vaccine. Aust. Vet. J. 72:296-300. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

