Characterization of retinaldehyde dehydrogenase 3
- PMID: 16241904
- PMCID: PMC1386004
- DOI: 10.1042/BJ20050918
Characterization of retinaldehyde dehydrogenase 3
Abstract
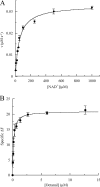
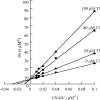
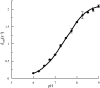
RALDH3 (retinal dehydrogenase 3) was characterized by kinetic and binding studies, protein engineering, homology modelling, ligand docking and electrostatic-potential calculations. The major recognition determinant of an RALDH3 substrate was shown to be an eight-carbon chain bonded to the aldehyde group whose kinetic influence (kcat/Km at pH 8.5) decreases when shortened or lengthened. Surprisingly, the b-ionone ring of all-trans-retinal is not a major recognition site. The dissociation constants (Kd) of the complexes of RALDH3 with octanal, NAD+ and NADH were determined by intrinsic tryptophan fluorescence. The similarity of the Kd values for the complexes with NAD+ and with octanal suggests a random kinetic mechanism for RALDH3, in contrast with the ordered sequential mechanism often associated with aldehyde dehydrogenase enzymes. Inhibition of RALDH3 by tri-iodothyronine binding in competition with NAD+, predicted by the modelling, was established kinetically and by immunoprecipitation. Mechanistic implications of the kinetically influential ionizations with macroscopic pKa values of 5.0 and 7.5 revealed by the pH-dependence of kcat are discussed. Analogies with data for non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans, together with the present modelled structure of the thioacyl RALDH3, suggest (a) that kcat characterizes deacylation of this intermediate for specific substrates and (b) the assignment of the pKa of the major ionization (approximating to 7.5) to the perturbed carboxy group of Glu280 whose conjugate base is envisaged as supplying general base catalysis to attack of a water molecule. The macroscopic pKa of the minor ionization (5.0) is considered to approximate to that of the carboxy group of Glu488.
Figures


Similar articles
-
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and phosphorylating glyceraldehyde-3-phosphate dehydrogenase.J Biol Chem. 1997 Jun 13;272(24):15106-12. doi: 10.1074/jbc.272.24.15106. J Biol Chem. 1997. PMID: 9182530
-
Kinetic characterization of recombinant mouse retinal dehydrogenase types 3 and 4 for retinal substrates.Biochim Biophys Acta. 2009 Dec;1790(12):1660-4. doi: 10.1016/j.bbagen.2009.09.004. Epub 2009 Sep 18. Biochim Biophys Acta. 2009. PMID: 19766701
-
Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.J Mol Biol. 1999 Jul 2;290(1):161-73. doi: 10.1006/jmbi.1999.2853. J Mol Biol. 1999. PMID: 10388564
-
Chemical mechanism and substrate binding sites of NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.Chem Biol Interact. 2001 Jan 30;130-132(1-3):15-28. doi: 10.1016/s0009-2797(00)00218-0. Chem Biol Interact. 2001. PMID: 11306027
-
The non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase: biochemistry, structure, occurrence and evolution.Biol Chem. 1997 Dec;378(12):1413-9. Biol Chem. 1997. PMID: 9461340 Review.
Cited by
-
Pharmacological inhibition of ALDH1A in mice decreases all-trans retinoic acid concentrations in a tissue specific manner.Biochem Pharmacol. 2015 Jun 1;95(3):177-92. doi: 10.1016/j.bcp.2015.03.001. Epub 2015 Mar 9. Biochem Pharmacol. 2015. PMID: 25764981 Free PMC article.
-
Structural shifts of aldehyde dehydrogenase enzymes were instrumental for the early evolution of retinoid-dependent axial patterning in metazoans.Proc Natl Acad Sci U S A. 2011 Jan 4;108(1):226-31. doi: 10.1073/pnas.1011223108. Epub 2010 Dec 17. Proc Natl Acad Sci U S A. 2011. PMID: 21169504 Free PMC article.
-
ALDH1A3 mutations cause recessive anophthalmia and microphthalmia.Am J Hum Genet. 2013 Feb 7;92(2):265-70. doi: 10.1016/j.ajhg.2012.12.003. Epub 2013 Jan 9. Am J Hum Genet. 2013. PMID: 23312594 Free PMC article.
-
Functions of Intracellular Retinoid Binding-Proteins.Subcell Biochem. 2016;81:21-76. doi: 10.1007/978-94-024-0945-1_2. Subcell Biochem. 2016. PMID: 27830500 Free PMC article. Review.
-
The stem cell/cancer stem cell marker ALDH1A3 regulates the expression of the survival factor tissue transglutaminase, in mesenchymal glioma stem cells.Oncotarget. 2017 Apr 4;8(14):22325-22343. doi: 10.18632/oncotarget.16479. Oncotarget. 2017. PMID: 28423611 Free PMC article.
References
-
- Gagnon I., Duester G., Bhat P. V. Enzymatic characterization of recombinant mouse retinal dehydrogenase type. Biochem. Pharmacol. 2003;65:1685–1690. - PubMed
-
- Wang X., Penzes P., Napoli J. L. Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. Recognition of retinal as substrate. J. Biol. Chem. 1996;271:16288–16293. - PubMed
-
- Grün F., Hirose Y., Kawauchi S., Ogura T., Umesono K. Aldehyde dehydrogenase 6, a cytosolic retinaldehyde dehydrogenase prominently expressed in sensory neuroepithelia during development. J. Biol. Chem. 2000;275:41210–41218. - PubMed
-
- Lin M., Zhang M., Abraham M., Smith S. M., Napoli J. L. Mouse RALDH4: molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J. Biol. Chem. 2003;278:9856–9861. - PubMed
-
- Li H., Wagner E., McCaffery P., Smith D., Andreadis A., Dräger U. C. A retinoic acid synthesizing enzyme in ventral retina and telencephalon of the embryonic mouse. Mech. Dev. 2000;95:283–289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

